Designing disease-resistant crops
From basic knowledge to biotechnology

Ancient records describe how plant diseases were attributed to many causes, including divine power, religious belief, and superstition. Far from these days, we now have detailed knowledge about how plant immunity is executed. Plants employ two types of sensors to perceive and defeat the litany of pathogenic organisms that attack them, whilst microbes deploy a myriad of specialized weapons to suppress immunity and promote infection. This opens a path to exploiting these insights to increase crop resistance. Here we describe novel biotechnological approaches for designing superior disease-resistant crops to fight agricultural losses in the field while reducing chemical inputs, towards a more sustainable agriculture that ensures food security.
Keywords: crops, diseases, biotechnology, food security.
Life in the twenty-first century makes it difficult to appreciate how plant diseases have impacted on human societies and our own history. In one devastating example, the Irish Potato Famine of 1845 occurred when the late blight disease of potato caused by the oomycete pathogen Phytophthora infestans (Figure 1) became an uncontrollable epidemic that overwhelmed society. This resulted in the death of roughly one million people from starvation, while a further million were forced to emigrate, giving rise to the famous Irish diaspora. Hundreds of years later, potato late blight remains arguably the most damaging potato pathogen worldwide. According to the Food and Agriculture Organization of the United Nations (FAO), between 20 to 40 % of worldwide production for major crops is lost to diseases and pests each year (Oerke, 2006). Not surprisingly, crop losses are highest in those regions that are already facing hunger and food insecurity. The world’s population is projected to reach 9.1 billion by 2050, 34 % higher than today. Thus, modern agriculture must provide for the world’s increasing population and protect it from uncontrollable epidemics that threaten food security.
«Current crops are the product of thousands of years of human selection and classical breeding from their original, wild forms»
Currently, food production relies on agricultural practices that include cultural and biological controls, deployment of agrochemicals, and the intentional use of resistant crop varieties. While pesticides have contributed much to our food security, the regular use of agrochemicals raises significant concerns over their negative health and environmental effects, highlighting the need to transition to a more sustainable agriculture. The intentional use of host plant resistance is an ecologically benign and economically efficient means of managing crop diseases. Humans have selected disease resistant plants since the dawn of agriculture. For modern breeders, the development of a disease resistant cultivar contains two steps. Firstly, the identification of resistant plants, normally from closely-related wild cultivars. And secondly, the incorporation of the immune genes responsible for disease resistance into selected crops by classical breeding or genetic engineering. Classical breeding methodologies introgress resistance genes into elite crop cultivars by crossing and selecting the offspring for the desired traits. In fact, current crops are the product of thousands of years of human selection and classical breeding from their original, wild forms. This is a lengthy and labour-intensive process that restricts genetic crop improvement within those species that are sex-compatible.

Figure 1. The pathogen Phytophthora infestans was the cause of the late nineteenth-century potato late blight epidemic in Ireland, which led to what is commonly known as the Great Irish Famine of 1845. In the pictures, effects of the pathogen on a potato and on the leaves of the potato plant. / Howard F. Schwartz, Colorado State University (CC BY) / Ben Millet CC BY-NC-ND 2.0
An alternative way to introduce new resistance genes into elite crop cultivars is by using genetic engineering, which allows the direct alteration of an organism’s genes using biotechnology. This allows the insertion, modification or deletion of selected genes. Perhaps the most important advantage of genetic engineering is that it allows the interchange of genetic material across species barriers, or the introduction of new resistance genes into vegetatively propagated (non-sexual) staple crops such as banana, cassava, and potato, overcoming the limits imposed by classical breeding. Genetic engineering provides the potential to greatly expand the efficiency and precision of possible modifications by the use of cutting-edge genome-editing technologies such as CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated endonuclease 9). Crop cultivars improved by such methods are known as genetically modified crops. All of these genetic engineering strategies expand the possibilities for introducing disease-resistance into crops where classical breeding programs are challenging or even not feasible.
The plant immune system
Research over the last 30 years in model and crop plant species has led to an increasingly detailed conceptual understanding of the molecular mechanisms controlling plant disease resistance and susceptibility, elegantly synthesised by Jones and Dangl in the so-called «zigzag model» (Jones & Dangl, 2006). This theory postulates that plants employ two types of sensors to resist the attack of pathogenic organisms, while microbes promote infection by using a battery of specialized weapons. On one side, the frontline of plant defense is provided by specialized cell surface immune sensors that detect the microbial presence outside of the plant cell through perception of conserved microbial structures or patterns. This recognition activates plant immunity, and is the reason why plants are generally resistant to the vast number of microbes that surround them. How is it then possible that plant pathogens exist? In response, sucessful pathogens produce phytotoxins and deliver virulence molecules called «effectors» into plant cells to attack plant host targets and promote infection. Effectors contribute collectively to pathogenesis by attacking multiple cellular defensive processes and their action is essential for disease progression. The second line of plant defense is performed by intracellular immune receptors that detect effectors upon their injection into plant cells. After perception, intracellular receptors execute immunity in its strongest form, which is characteristically associated with host cell death at the infection site, restoring full immunity. This «zigzag model» predicts an ongoing co-evolutionary battle between plants and their pathogens that drives adaptive evolutionary change to shift the balance of power towards either partner, but also provides insights that can be used to engineer disease-resistant crops.
Recruiting cell surface sensors to the frontline of defenses
A promising new strategy for increasing crop disease resistance is based on the deployment of specialized cell surface immune sensors to improve pathogen recognition. Cell surface sensors provide quantitatively enhanced resistance to diseases. By definition, microbial patterns perceived by these sensors are molecular components that are highly conserved within a class of microbes and with an essential function for fitness or survival, which implies that they cannot be easily modified by microorganims to overcome their perception. For example, chitin, a major structural element of all fungal cell walls, acts as an important microbial pattern that is perceived by most plant species, activating plant immunity. Thus, one way to achieve broader spectrum resistance that might also be durable is to transfer these specialized cell surface sensors that detect common microbial molecules of a broad class of microbes, such as chitin for fungal pathogens, into those crops that lack them.

Figure 2. Genetic engineering strategies for transferring a series of cell surface sensors to crop species have been shown to be very effective. Among others, these strategies have provided increased resistance to diseases such as the bacterial wilt of the banana caused by Xanthomonas, which poses a threat to agricultural production in some regions. In the pictures at the top, details from fruits and leaves affected by Xanthomonas campestris, and below, banana plantation in Tanzania, showing the effects of the bacterium. / IITA / FAO – Fidelis Kahiura
In 2010, a major breakthrough came when the group of Cyril Zipfel showed that transfer of the cell surface sensor perceiving the bacterial elongation factor Tu, one of the most abundant microbial patterns in bacteria, from a small plant in the mustard family to tomato using genetic engineering techniques, increased resistance to a wide range of bacterial pathogens (Lacombe et al., 2010). Since then, similar genetic engineering approaches have been used to transfer a number of cell surface sensors across family boundaries into crop species (Rodriguez-Moreno et al., 2017). Globally, these strategies have proved to be highly effective, providing quantitatively enhanced resistance to diseases such as citrus canker of sweet orange, bacterial leaf blight of rice, bacterial halo blight of wheat, late blight of potato, and Xanthomonas bacterial wilt of banana, the latter of which is one of the most important threats to banana production in the Great Lakes region of Africa (Figure 2). Thus, cell surface immune sensors represent a potent source of long-lasting resistance that can be transferred even between very distantly related plant families for disease control. Plant genomes encode several hundreds of these potential sensors that may have roles in microbe perception. Nowadays, identification of novel cell surface immune sensors that perceive specific microorganisms and their transfer to crops is an active area of research that emerges as an exciting approach to improve crop resistance to important pathogens.
Creating intracellular receptor traps against critical diseases
Intracellular immune receptors are important tools in breeding programs for managing crop diseases. This type of immunity relies on a highly specific recognition mechanism that triggers resistance when an intracellular immune receptor recognizes into the plant cell its corresponding effector, that has been injected by an approaching pathogen to induce disease. Thus, intracellular receptors typically confer resistance to only those races of the pathogen that contain the specific effector that is recognized, but which is in turn, more rapid, intense and effective against the appropriate pathogen by comparison to resistance triggered by cell surface sensors. Commonly, intracellular receptors provide full resistance to an specific pathogen, while cell surface sensors contribute quantitatively to resistance to a broad class of microbes.
The effectiveness of this type of resistance was first demonstrated by Sir Rowland Biffen in his wheat disease-resistance breeding program in the early twentieth century (Biffen, 1905). Since then, intracellular immune receptors have been widely deployed, particularly through introduction of such genes into classical breeding programs from resistant wild relatives. Concomitantly, numerous intracellular receptors have been identified and introduced into plants through genetic engineering techniques to generate varieties resistant to important diseases. In a prominent example, introduction of an intracellular immune receptor known as Rpi-vnt1.1, isolated from a wild South American potato relative, into cultivated potato fully protected it from the devastating late blight (Figure 3) (Foster et al., 2009). Remarkably, a late blight resistant potato variety containing this intracellular receptor became in 2015 the first genetically modified crop with enhanced resistance towards a non-viral pathogen to be approved for commercial use in the USA. Nowadays, finding novel intracellular immune receptors is an extremely active area of research for safeguarding global crops against their most devastating pathogens.
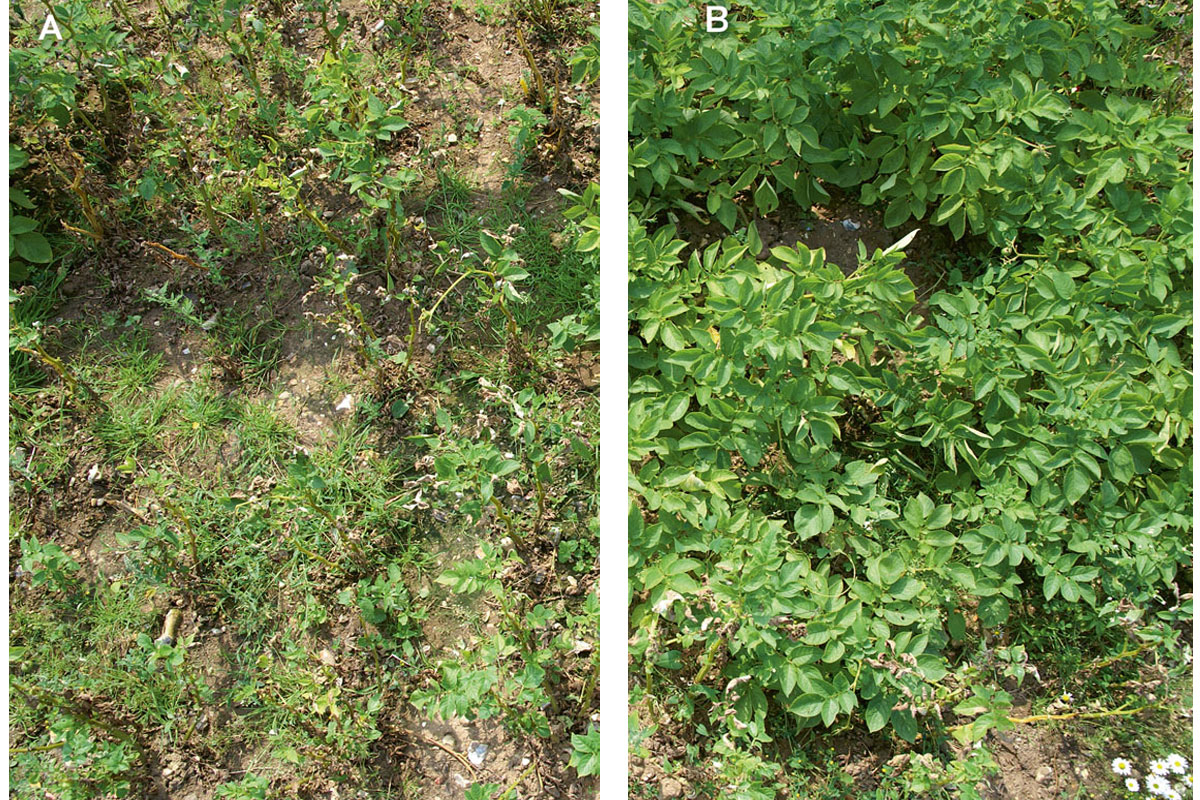
Figure 3. In the pictures, field trials at an advanced stage of late blight outbreak. They show the original potato cultivated variety Desiree (A) and the same variety containing the intracellular receptor Rpi-vnt1.1 (B), with no evident symptoms of potato late blight, caused by the pathogen Phytophthora infestans. Images were taken in 2012 field trials and were kindly provided by Prof. Jonathan D. Jones (The Sainsbury Laboratory, Norwich, UK).
Intracellular immune receptors have been transferred between closely related plant species with relatively good success. In contrast, their introduction into more distant plants, such as distinct plant families, commonly fails. This phenomenon has come to be known as restricted taxonomic functionality and is the main reason why researchers prefer to identify intracellular immune receptors from wild related species of crops, which are more likely to function once introduced into a closely current cultivated plant. An alternative way to overcome this problem might be through the direct redesign of endogenous immune receptors of a crop to expand their recognition capabilities. In this regard, a synthetically modified version of a potato intracellular immune receptor known as R3a was recently generated, extending its pathogen recognition capabilities towards other effectors from the oomycete pathogen Phytophthora infestans (Segretin et al., 2014), whereas another intracellular receptor from tomato known as I2 has already been engineered to confer simultaneously partial resistance to Phytophthora infestans in addition to the fungus Fusarium oxysporum (Giannakopoulou et al., 2015). Although these experiments were performed in a model plant with no agricultural value, this pioneering work suggests that endogenous immune receptors can be directly engineered to confer resistance to diverse pathogens. These insights may also be extended using novel genome-editing technologies such as CRISPR/Cas9 which offer precise modification of existing host genes, which could be exploited to develop new recognition specificities in pre-existing intracellular immune receptors of major crops.
Protecting plants’ Achilles’ heel
Plant pathogens use a variety of molecular strategies to quell host immunity. The host targets of these strategies are commonly known as plant susceptibility genes, because their manipulation is required for disease progression. Remarkably, multiple examples support common evolution by unrelated pathogens to attack common plant susceptibility targets, which designate these hubs as a major Achilles’ heel for the plant. Thus, a recent strategy for protecting crops against diseases is based on the removal or modification of such susceptibility genes to avoid their manipulation by pathogens.
«Genetic engineering approaches are emerging as a feasible, specific, and versatile strategy for bolstering disease resistance in crops»
A number of recent examples illustrate how novel techniques for genome editing can be successfully applied in crops to protect plant susceptibility genes that are attacked by pathogens. Tactics frequently employed by plant pathogens to promote disease involve for example manipulation of defensive genes activated during the infective process and hijacking of phytohormone pathways, which include the two main defensive pathways in plants based on the salicylic acid and jasmonic acid hormones. In this context, a tomato jasmonic acid co-receptor called JAZ2 was recently re-edited by using CRISPR/Cas9 technology to prevent its manipulation by a phytotoxin produced by multiple strains of the bacterium Pseudomonas syringae to promote disease (Figure 4). This resulted in a tomato line that was resistant to bacterial speck disease caused by this phytopathogenic bacterium (Ortigosa et al., 2018). In another very elegant and recent example, a similar strategy was used to modify three susceptibility genes in rice, known as SWEET genes, which has allowed to engineer broad and durable resistance to bacterial blight caused by the bacterial pathogen Xanthomonas oryzae in rice, a serious disease in much of Asia and parts of Africa (Oliva et al., 2019). All of these examples highlight how new genome editing technologies can be used to deliver resistance to crops that could be readily implemented in the field.
Improved practices for breeding durable resistance
Unfortunately, although very effective, disease resistance conferred by individual intracellular immune receptors is usually short-lived in the field. This is because pathogens can rapidly evolve to evade recognition, for example, by losing or modifying one recognised effector among their redundant effector repertoire. To overcome this problem, one strategy is to deploy multiline varieties that contain mixed seed lines with different single intracellular receptor genes, which reduces the selective pressure on a pathogen compared to resistant monocultures, which contain a single common intracellular receptor in each seed. Another strategy relies on the deployment of multiple intracellular immune receptors simultaneously in a single seed line, which is commonly known as «gene stacking» or «pyramiding». This strategy is predicted to provide long-lasting immunity, as multiple concomitant changes are required to overcome such resistance, which has an extremely low collective probability. The effectiveness of gene stacking is exemplified by a recent example in which stacking of three late blight intracellular immune receptors into African highland potato varieties by genetic engineering conferred complete field resistance to this disease, resulting in a three to four-fold productive increase over the national average (Ghislain et al., 2018). These late blight resistant potato varieties can be rapidly adopted given a supportive regulatory environment and bring significant income increases to smallholder farmers, whereas this disease causes annual yield losses from 15–30 % in sub-Saharan Africa.
«Nowadays, there are only a few examples of genetically engineered disease-resistant crops that have made it to the commercial level»
Although most previous strategies were focussed on deployment of individual intracellular immune receptors with good success, it is likely that the most effective strategy to combat diseases durably in the field is still to come. This must be built on a multi-layered pyramiding of defences, where the same cultivar could combine cell surface sensors, intracellular immune receptors, and disabling of specific susceptibility genes. Together, these modifications would bring potential pathogens to their knees. This should confer increased long-lasting resistance to a wide-range of pathogens through cell surface sensors and full immunity to specific pathogenic strains with intracellular immune receptors and disabled susceptibility genes, which might be even more durable in the field.
The future that awaits
Genetic engineering approaches are emerging as a feasible, specific, and versatile strategy for bolstering disease resistance in crops, especially in those vegetatively propagated crops in which classical breeding programs are precluded. The directed methods described here could enable the development of «improved» crops that sense and respond more effectively to invading pathogens, or avoid the ways in which microbes twist crops around their little fingers, potentially without reductions in crop productivity. Nowadays, there are only a few examples of genetically engineered disease-resistant crops that have made it to the commercial level, because of the currently restrictive legislation around genetically modified crops, and despite the scientific consensus is that genetically engineered crops are as safe as those developed by classical methods (National Academies of Sciences, Engineering, and Medicine, 2016). These biotechnological strategies are more effective, environmentally friendly, and safer than many current methods of control based on agrochemical use. Indeed, functional intracellular immune receptor gene stacking in late blight resistant potato is estimated to reduce fungicide use by over 80 % (Haverkort et al., 2016). As the world’s population continues to grow, we cannot continue to ignore genetic engineering aproaches for delivering host resistance into crops. The reason is simple: these superior disease-resistant varieties hold the potential to fight agricultural losses to pests while reducing chemical inputs, and promise a more sustainable agriculture that ensures food security.
References
Biffen, R. H. (1905). Mendel’s laws of inheritance and wheat breeding. The Journal of Agricultural Science, 1(1), 4–48. http://doi.org/10.1017/S0021859600000137
Foster, S. J., Park, T. H., Pel, M., Brigneti, G., Sliwka, J., Jagger, L., van der Vossen, E., & Jones, J. D. G. (2009). Rpi-vnt1.1, a Tm-2(2) homolog from Solanum venturii, confers resistance to potato late blight. Molecular Plant-Microbe Interactions, 22(5), 589–600. http://doi.org/10.1094/MPMI-22-5-0589
Ghislain, M., Byarugaba, A. A., Magembe, E., Njoroge, A., Rivera, C., Román, M. L., Tovar, J. C., Gamboa, S., Forbes, G. A., Kreuze, J. F., Barekye, A., & Kiggundu, A. (2018)). Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnology Journal, 17(6), 1119–1129. http://doi.org/10.1111/pbi.13042
Giannakopoulou, A., Steele, J. F. C., Segretin, M. E., Bozkurt, T. O., Zhou, J., Robatzek, S., Banfield, M. J., Pais, M., & Kamoun, S. (2015). Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Molecular Plant-Microbe Interactions, 28(12), 1316–1329. http://doi.org/10.1094/MPMI-07-15-0147-R
Haverkort, A. J., Boonekamp, P. M., Hutten, R., Jacobsen, E., Lotz, L. A. P., Kessel, G. J. T., Vossen, J. H., & Visser, R. G. F. (2016). Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: Scientific and societal advances in the DuRPh project. Potato Research, 59(1), 35–66. http://doi.org/10.1007/s11540-015-9312-6
Jones, J. D., & Dangl, J. L. (2006). The plant immune system. Nature, 444(7117), 323–329. http://doi.org/10.1038/nature05286
Lacombe, S., Rougon-Cardoso, A., Sherwood, E., Peeters, N., Dahlbeck, D., van Esse, H. P., Smoker, M., Rallapalli, G., Thomma, B. P. H. J., Staskawicz, B., Jones, J. D. G., & Zipfel, C. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nature Biotechnology, 28(4), 365–369. http://doi.org/10.1038/nbt.1613
National Academies of Sciences, Engineering and Medicine. (2016). Genetically engineered crops: Experiences and prospects. The National Academies Press. http://doi.org/10.17226/23395
Oerke, E. (2006). Crop losses to pests. The Journal of Agricultural Science, 144(1), 31–34. http://doi.org/10.1017/S0021859605005708
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., Eom, J.-S., Li, C., Nguyen, H., Liu, B., Auguy, F., Sciallano, C., Luu, V. T., Dossa, G. S., Cunnac, S., Schmidt, S. M., Slamet-Loedin, I. H., Vera Cruz, C., Szurek, B., … Yang, B. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nature Biotechnology, 37(11), 1344–1350. http://doi.org/10.1038/s41587-019-0267-z
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., & Solano, R. (2018). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnology Journal. http://doi.org/10.1111/pbi.13006
Rodriguez-Moreno, L., Song, Y., & Thomma, B. P. (2017). Transfer and engineering of immune receptors to improve recognition capacities in crops. Current Opinion in Plant Biology, 38, 42–49. http://doi.org/10.1016/j.pbi.2017.04.010
Segretin, M. E., Pais, M., Franceschetti, M., Chaparro-Garcia, A., Bos, J. I. B., Banfield, M. J., & Kamoun, S. (2014). Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Molecular Plant-Microbe Interaction, 27(7), 624–637. http://doi.org/10.1094/MPMI-02-14-0040-R