Next Stop: Language
The ‘FOXP2’ gene’s journey through time

How did humans evolve language? The fossil record does not yield enough evidence to reconstruct its evolution and animals do not talk. But as the neural and molecular substrates of language are uncovered, their genesis and function can be addressed comparatively in other species. FOXP2 is such a case – a gene with a strong link to language that is also essential for learning in mice, birds and even flies. Comparing the role FOXP2 plays in humans and other animals is starting to reveal common principles that may have provided building blocks for language evolution.
Keywords: speech, language, sensory-motor learning, evo-devo, deep homology.

Figure 1. What would be most interesting for an extraterrestrial scientist who lands on planet Earth? / Peter Schatton
Lenguage is universal
Imagine yourself as an extraterrestrial behavioral scientist that lands for the first time on our planet (Figure 1). Would you marvel most at the gigantic whales and tiny fruit-flies going about their business of foraging and reproducing in very different ways? Or would you be more fascinated by those bi-pedally walking cloth-wearing apes that inhabit vastly different habitats across the globe, looking quite diverse and chattering away at each other with very different sound streams, but all belonging to the same species, Homo sapiens? Looking closer, you will find that these different sound streams, what we call languages, are in fact fundamentally universal – every human society has its own language.
While sounding different, all languages share some commonalities. All languages are hierarchically structured. Their building blocks are sounds that humans can produce effortlessly with their voice, such as p, b, f, v, t, d, a, e and so on. They are combined into a larger repertoire of syllables out of which thousands of different words can be composed. Rules of grammar that are also partially similar across all languages allow for the stringing together of words into almost limitless ways to build sentences that can express seemingly unlimited facts and thoughts. The same holds true for sign language, where a limited repertoire of hand and face movements is combined into a larger repertoire of gestures and those, using grammar, and used to form sentences that convey meaning.
Our extraterrestrial behavioral scientist might think: «Very nifty system», but why do such different languages share some common principles?
«Beginning a few weeks after birth, infants can distinguish speech sounds from other sounds and even match a syllable they hear to the face that makes that sound»
Genetic contributions to language
The fact that all humans use language, and that all languages share some characteristics, have suggested to linguists and biologists that there must be a strong genetic component. The genetic component of language, called «the language instinct» in the title of Steven Pinker’s book published in 1994, must have something to do with the ability to «learn» language because the genetic makeup of a baby has nothing to do with which particular language it will eventually speak; a baby born to Spanish-speaking parents but adopted and raised by French-speaking parents will learn French, not Spanish. So how does this learning process take place? First, the baby’s brain needs to identify what language sounds are. Beginning a few weeks after birth, infants can distinguish speech sounds from other sounds and even match a syllable they hear to the face that makes that sound (Kuhl & Meltzoff, 1982). When a baby starts babbling at a few months of age, it has to modify the babbling sounds towards the language of its environment. This process continues through the first two years of life. In parallel, the baby learns – effortlessly and without formal instruction – the meaning of the sounds and the norms that rule how these are put together. Even before toddlers can speak well, they know the difference in meaning when parents ask «Did you take Mary’s toy?» or «Did Mary take your toy?». As children get older their brains also react differently to sentences that are semantically correct or incorrect («my uncle will watch the movie» versus «my uncle will blow the movie») and also differentiate between syntactically correct and incorrect sentences («My uncle will watch/watching the movie») (Friederici, 2006).
«Wow – says the extraterrestrial –, these Homo sapiens babies seem to be born as little language learning machines. How can they do this?»
FOXP21
If babies are born with an instinct for learning language, how is this anchored in their genes? Does it require «specialized» genes that are only relevant for language and speech, or is language learning just another learning process, like learning to walk, to ride a bicycle or to do math? The latter seemed to be more plausible as genetic diseases affecting language usually also impair other cognitive domains. Thus it came as a surprise to the scientific community when multiple members of a family in Great Britain were diagnosed with a «specific language impairment» in the absence of other morphological or mental abnormalities. The grandmother of the so-called KE family apparently had a spontaneous mutation that was inherited by four of her five children and ten of her 24 grandchildren, affecting both boys and girls. This inheritance pattern is characteristic of a dominant mutation on one of the non-sex chromosomes (autosomes). In the case of the KE family, the affected family members have problems with the coordination of the tongue and mouth muscles to orchestrate speech sounds. For example, repeating nonsense multisyllable words like «woogalamic» or «perplisteronk» is much more difficult for a KE family member who has the mutation than for a non-affected sibling (Watkins, Dronkers, & Vargha-Khadem, 2002). In addition, grammatical comprehension is also affected, but less so than speech production. For instance, when asked to point to the appropriate picture for «the hare chasing the horse is red», the affected family members perform worse (Figure 2).
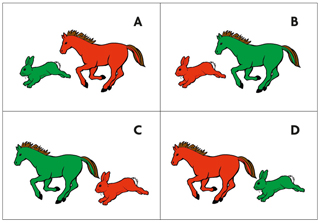
Figure 2. Image designed based on TROG-test (Test of reception of grammar, Bishop, 1983). Affected KE family members perform worse when being asked to point to the appropriate picture for: «the hare chasing the horse is red». / Adriana Schatton
Since problems understanding and producing complex language impact education and interaction with others, the mutation represents a huge challenge for the affected family members. At the same time, it provided the scientific community with a window into the molecular underpinnings of language production and perception, and raised hope for the possibility of clinical intervention (Vargha-Khadem, Gadian, Copp, & Mishkin, 2005). Genetic mapping revealed that all the affected KE family members had a mutation in a gene on chromosome 7 called Forkhead box P2, or FOXP2 (Lai, Fisher, Hurst, Vargha-Khadem, & Monaco, 2001). The amusing name forkhead box was coined in 1989 by scientists working on fruit fly development who identified mutants with a «forked head» and linked this phenotype to a particular stretch of DNA, called box. This «forkhead box» exists in more than forty genes that are organized into nineteen gene families named FoxA through FoxS. They code for proteins that regulate the function of other genes, so called «transcription factors». The «forkhead box» part of the transcription factor binds to specific regulatory regions of other genes, so called «target genes», leading to their reduced or enhanced activity. Many forkhead transcription factors play roles in health and disease but only the P gene family has been linked to speech and language.
The extraterrestrial scientist is scratching its own «forkhead» antennae and wondering: «If I had a FOXP2 gene, would I be able to speak like those Homo sapiens do?»
«The discovery of FOXP2 opened the door for answering many questions about the link between genes and language»
Unique genes for unique traits?
The discovery of FOXP2 opened the door for answering many questions about the link between genes and language. Does having a FOXP2 gene give you language? No. FoxP2 exists in many species that do not talk. The importance of this gene for lung development had been described in mice even before its mutation in the KE family was linked to language, showing that FoxP2 was neither a gene unique to humans nor unique to the brain. Human-unique genes do exist (Dennis et al., 2012) but FoxP2 is not one of them. FoxP2 exist in all 274 vertebrates studied so far, and it is also among the genes whose sequence barely changed throughout vertebrate evolution. This suggests that the function of FoxP2 may not have changed much from fish to humans, so that whatever the exact function of this gene may be, there is evolutionary pressure for conserving the FoxP2 sequence. This is supported by the findings that FOXP2 mutations identified in humans – and there are now 23 different ones, all impairing speech and language – never affect both gene copies (alleles), implying that when both copies are mutated the embryo cannot survive. This is also the case in mice that carry mutations on both Foxp2 alleles: they die a few weeks after birth, apparently of lung disorders. An exception from this high conservation for FoxP2 in vertebrates are bats. Many bat species have a widely diverged FoxP2 sequence, which is an enigmatic finding waiting for an explanation (for review see Knörnschild, 2014).
The extraterrestrial may find this a bit confusing. FOXP2 mutations cause a deficit in a uniquely human trait, language, but FoxP2 exists in all vertebrates in pretty much the same version?
Of mice and men
Just as it seemed that FOXP2 was not so special after all, Wolfgang Endard and colleagues at the Max Planck Institute for Evolutionary Anthropology in Leipzig reported that the sequence of the FOXP2 protein in humans, while overall very similar to all vertebrate FoxP2s, had in fact changed more in (evolutionary) recent history than expected. Moreover, all humans carry this specifically human FOXP2 version starting about 200,000 years ago, roughly coincident with the emergence of anatomically modern humans, even though the exact time is controversial. The notion that the human-unique version of this protein was important for human evolution was met with skepticism because the sequence differed from the chimp version in only two of 714 amino acids, the building blocks of proteins. Could just two amino acids be the reason why we speak and chimps do not? On the other hand, the discovery of a human-specific FOXP2 version that was apparently beneficial to carry and thus swept through the entire human population begged further investigation. The Leipzig scientists therefore designed an ingenious experiment: they experimentally added the human FOXP2 gene to mice and checked whether those mice were different from their non-manipulated siblings (Enard et al., 2009). Not surprisingly, the mice did not develop language, establishing that the human FOXP2 version alone cannot kick-start all the changes in the brain and the vocal apparatus required to develop speech and language.
«When mice pups carrying the additional humanized FOXP2 version were separated from their mother they made distress calls that had different bioacoustics features than calls of their non-manipulated siblings»
Given the differences of mouse and human brains and sound producing structures, it was of course highly unlikely that a single gene could trigger such a program. However, the experiment introducing the human FOXP2 gene into a mouse was informative; when mice pups carrying the additional humanized FOXP2 version were separated from their mother they made distress calls that had different bioacoustic features than calls of their non-manipulated siblings, providing another link of FOXP2 to vocalizations. The neural substrates for these vocalization changes are not known yet, but a number of different follow up experiments showed that mouse brains exposed to a humanized version of FOXP2 differ structurally and functionally from brains of mice with mouse-Foxp2. Additional research from the labs of Ann Graybiel, Simon Fisher and Wolfgang Enard established that brains of yet a different mouse line that was experimentally altered to carry an equivalent mutation to the one of the KE family also show differences to wild-type mice. Interestingly, it seems that in mice some of the neural and behavioral consequences of having a Foxp2 allele with a mutation similar to the one that produces language impairments in humans are the opposite of the consequences of carrying an additional humanized version of the gene. The latter also differ from wild type litter mates in a maze learning task. When mice have to switch between finding their way based on visual cues (e.g., turn to where you see a star) to finding their way by turning left or right, the humanized-FoxP mice show a quicker transition from one strategy to the other. The authors suggest that language learning requires a similar type of switch in learning styles.
Together, these findings highlight that small sequence changes in a highly conserved gene might change neural function but that unique behavioral traits or morphological adaptations do not require unique genes. Rather, particular combinations of sets of genes can act together causing «unique» effects. In fact, since the original discovery of the link between FOXP2 mutations and the specific language impairment, more genes implicated in language-related disorders have been identified. Some of these are likely to act together with FOXP2 in a molecular network. For instance, another FoxP gene family member, FOXP1, is also linked to language deficits and the FoxP1 and FoxP2 proteins can directly interact (Graham & Fisher, 2015).
The extraterrestrial scientist shouts: «What a funky idea introducing the human version of a gene into mice!» And goes on to wonder why those mice had neural and behavioral changes, even if they did not start to speak…
Formation of circuits versus function in circuits
Did the human version of FOXP2 in mice affect brain development? Or did the gene manipulation affect the function of the brain after development was completed? Do FOXP2 mutations in humans hamper the embryonic development of brain circuits that children later need for language learning? Or does FOXP2’s activity play a role during the language learning process itself? The latter question cannot be answered in mice, because their vocalizations are, unlike human language, not learned by imitating the sounds of adult conspecifics. Children born deaf do not learn to speak as well as hearing children, because speech learning depends on auditory feedback. In contrast, deaf mice vocalize exactly like hearing mice demonstrating that mouse vocalizations are not imitatively learned.
Unlike mice, some whale and bat species do modify the sounds they use for communication as a result of experience (Janik & Slater, 1997; Knörnschild, 2014). These vocal learners are hard to keep in laboratories and consequently gene-function studies in bats and whales have not yet been performed. Many song bird species also learn to produce their songs by imitating adult conspecifics and this process bears many behavioral and neural similarities to speech learning (Bolhuis, Okanoya, & Scharff, 2010) (Figure 3). In addition, the neural structures underlying song learning and song production are known and the genomes of around fifty songbird species were recently sequenced (Zhang et al., 2014) making comparative neurogenetic studies increasingly attractive. In this vein, researchers in our group in Berlin, Germany, established that FoxP2 expression is modulated during the song learning period in a brain region essential for song learning (Haesler et al., 2004). Moreover, experimental reduction of FoxP2 levels in this brain area lead to incomplete and inaccurate song imitation in zebra finches (Haesler et al., 2007). Follow up experiments by us and other researchers have identified factors that regulate FoxP2 expression and cellular/behavioral consequences of its action (Condro & White, 2014; Wohlgemuth, Adam, & Scharff, 2014). These and other studies have firmly established that FoxP2 has two roles, one in neural development and an «on line» function in already developed neural circuits (Murugan, Harward, Scharff, & Mooney, 2013).
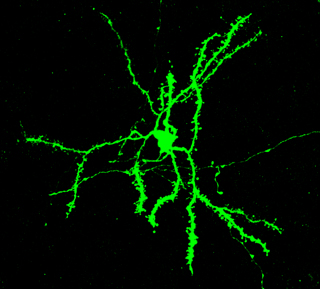
Figure 4. In songbirds and mammals, the FoxP2 protein is expressed in so called medium spiny neurons of the striatum, a region important for translating sensory information into motor behavior. These neurons are called spiny because they are studded with «spines», the sites where signals from other neurons are received. / Jennifer Kosubek
The extraterrestrial researcher recapitulates: «Songbirds learn to sing somewhat like humans learn to speak, and both need FoxP2 to get it right. How could that be?»
Common Themes
The fact that FoxP2 is important for both human speech and birdsong, two behaviors that have many similarities, as well as for motor learning in mice, is interesting from an evolutionary perspective. One common theme is that this transcription factor acts in neural circuits that are relevant for the translation of sensory stimuli (auditory in humans and birds, visual and tactile in mice) into motor acts (speech in humans, song in birds, and locomotion in mice). The neural circuits that are central for all these behavior involve the so called basal ganglia which are homologous in birds, mice and humans. «Homologous» in this context means that these structures seem to be derived from structures that already existed in a common ancestor of birds and mammals 300 million years ago. Form and function of the basal ganglia are strikingly similar in the avian and mammalian branches. One popular theory, supported by many empirical findings, postulates that sensory-motor learning in this circuit relies on a neurochemical reward system (e.g., dopamine) (Figure 4). Experimental evidence in mice and birds suggests that FoxP2 and its associated molecular network, including the two other neurally expressed FoxP proteins, FoxP1 and FoxP4, interact with dopamine signaling and regulate the strength of particular connections between particular sets of neurons, fine tuning sensory-motor integration (Wohlgemuth et al., 2014).
«The fact that FoxP2 is important for both human speech and birdsong, two behaviors that have many similarities, as well as for motor learning in mice, is interesting from an evolutionary perspective»
«Ah, I start to see the common thread running through this story», the extraterrestrial will hopefully say. FOXP2 together with other molecules seems to be active in parts of the brain that translate sensory experience into motor action. It may help a baby to modify the babbling sounds it makes to turn them into words, a young songbird to adjust its babbling to sound like his father’s song and a mouse to learn to use its body effectively to get food and not get eaten.
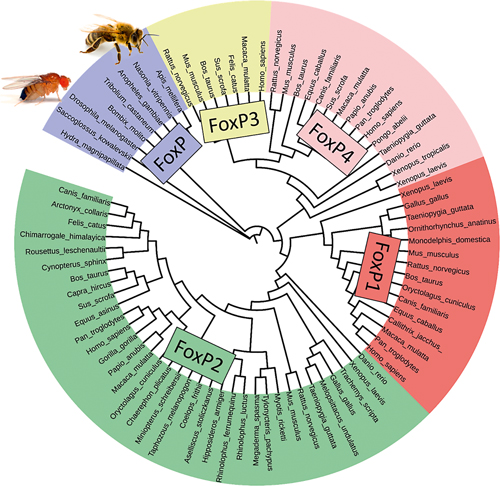
Figure 5. Invertebrate FoxP gave rise through gene duplication to four different FoxP genes. In vertebrates, FoxP1, 2, and 4 are important for brain development and behavior and play a role in other organs too. FoxP3 functions in the immune system. / Ezequiel Mendoza
Going back in time
Translating sensory experiences into motor action is a requirement for survival not exclusive to vertebrates. If FoxP2 is important for this function, did it exist before the vertebrate lineage arose? The gene locus FoxP2 that gave rise to the four different FoxP genes in vertebrates is indeed very old. It seems that these genes appeared together with animal multicellularity. Unicellular choanoflagellates do not have the gene but sponges and other nonbilaterians, e.g., animals without bilateral symmetry, do (Shimeld, Degnan, & Luke, 2010) (Figure 5). In sponges FoxP functions in cell adherence (on a plastic dish) and the formation of aggregates in epithelial-like layers. In the mouse lung, FoxP1 and FoxP4 are involved in the development of the lung epithelium (Li et al., 2012). Whether this is an «epithelial coincidence» or a case of deeper homology awaits further study. In the bilaterian lineage there is also only one FoxP gene, yielding two different versions, called isoforms. Interestingly for a possible similarity of function, in embryos and adults of the fruit fly (Drosophila melanogaster), FoxP is also expressed in the nervous system (Santos, Athanasiadis, Leitão, DuPasquier, & Sucena, 2011).
Turning to insects to pursue the idea that FoxP may fulfill similar functions in invertebrates and vertebrates, we and other groups have started to study the expression pattern and function in fruit flies and honeybees (Apis mellifera) (DasGupta, Ferreira, & Miesenböck, 2014; Kiya, Itoh, & Kubo, 2008; Lawton, Wassmer, & Deitcher, 2014; Mendoza et al., 2014) (Figure 6). As is often the case in the initial stages of scientific discovery of a new subject, some findings agree while other diverge. On the agreement side, all FoxP mutant flies show interesting behavioral deficits that are at first sight not incompatible with each other and could also be conceptually related to the behavioral deficits observed in vertebrates with reduced FoxP2 dosage. The authors of one study observed reduced courtship behavior (Lawton et al., 2014), in another study the flies were able to use visual cues to avoid punishment but were unable to use somatosensory information for the same task (Mendoza et al., 2014) and in a third study the mutant flies had trouble with decision making using olfactory cues in different concentrations and punishment as an incentive to learn (DasGupta et al., 2014). In this last experiment, the phenotype could be traced to a particular small set of neurons that had previously been described to be important for decision making and were now shown to also express FoxP.
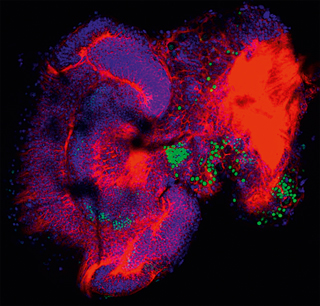
Figure 6. In the brain of bee larvae, FoxP is strongly expressed in different neuron clusters (green), indicating the importance of the gene for development.
Discrepancies exist with respect to the neural expression pattern of FoxP in Drosophila, even when almost identical approaches to visualize expression were used. Using green fluorescent protein to report FoxP promotor activity, Lawton et al. (2014) saw expression in the so called central complex, that has been hypothesized (Strausfeld & Hirth, 2013) to be functionally analogous to the vertebrate basal ganglia. In contrast, using a slightly different promoter sequence, DasGupta et al. (2014) did not find FoxP expression in the central complex at all but in the core region of the mushroom bodies instead, a multimodal integration site in the fly brain that is important for learning and memory. These differences might also account for the fact that in one study motor behavior (walking) was reported to be normal (DasGupta et al., 2014) whereas in other studies (Lawton et al., 2014) motor behavior was impaired. Needless to say, the devil with experimental conditions lies in the details and follow up studies using more approaches to determine where FoxP mRNA and protein is expressed in the nervous system of developing and adult flies as well as honey bees will refine the currently still confusingly complex results of the compare and contrast approach.

Figure 7. Our extraterrestrial scientist learned a lot about a gene and its journey through time. / Peter Schatton
Maybe our extraterrestrial observer would by now decide to stay a bit longer on this planet, roll up its sleeves and join the earthling scientists to figure out whether FoxP is indeed a candidate for «deep homology», fulfilling similar functions in neurons from flies to Homo sapiens. Or our space visitor may be tired from so much chitchat and jabbering all over the planet and yearning to go back to outer space, to think in silence (Figure 7).
Is language more than the sum of its parts?
Accompanying FoxP on its journey through evolutionary time is interesting as it points towards one building block of language learning: the translation of sensory information into motor acts. But language is of course more than that. Language allows people to speak truth or tell lies. Talk about yesterday’s events and tomorrow’s plans, abstract concepts like moral or logic, feelings such as happiness or sorrow. No genes have been directly linked to the process of how thoughts and feelings are externalized into speech. Maybe an extraterrestrial team of geneticists and linguists landing on Earth will write that next chapter on our path to understanding the evolution of language.
- By convention, when capitalized, FOXP2 refers to the human gene, Foxp2 refers to the gene in mice and FoxP2 refers to the gene in other species. The genes are italicized, but the proteins are not. (Back to the text)
REFERENCES
Bolhuis, J. J., Okanoya, K., & Scharff, C. (2010). Twitter evolution: Converging mechanisms in birdsong and human speech. Nature Reviews Neuroscience, 11, 747–759. doi: 10.1038/nrn2931
Condro, M. C., & White, S. A. (2014). Recent advances in the genetics of vocal learning. Comparative Cognition & Behavior Reviews, 9, 75–98. doi: 10.3819/CCB.2014.90003
DasGupta, S., Ferreira, C. H., & Miesenböck, G. (2014). FoxP influences the speed and accuracy of a perceptual decision in Drosophila. Science, 344, 901–904. doi: 10.1126/science.1252114
Dennis, M. Y., Nuttle, X., Sudmant, P. H., Antonacci, F., Graves, T. A., Nefedov, M., … Eichler, E. E. (2012). Human-specific evolution of novel SRGAP2 genes by incomplete segmental duplication. Cell, 149, 912–922. doi: 10.1016/j.cell.2012.03.033
Enard, W., Gehre, S., Hammerschmidt, K., Hölter, S. M., Blass, T., Somel, M., … Pääbo, S. (2009). A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell, 137, 961–971. doi: 10.1016/j.cell.2009.03.041
Friederici, A. D. (2006). The neural basis of language development and its impairment. Neuron, 52, 941–952. doi: 10.1016/j.neuron.2006.12.002
Graham, S. A., & Fisher, S. E. (2015). Understanding language from a genomic perspective. Annual Review of Genetics, 49, 131–160. doi: 10.1146/annurev-genet-120213-092236
Haesler, S., Rochefort, C., Georgi, B., Licznerski, P., Osten, P., & Scharff, C. (2007). Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus area X. PLoS Biology, 5, e321. doi: 10.1371/journal.pbio.0050321
Haesler, S., Wada, K., Nshdejan, A., Morrisey, E. E., Lints, T., Jarvis, E. D., & Scharff, C. (2004). FoxP2 expression in avian vocal learners and non-learners. The Journal of Neuroscience, 24, 3164–3175. doi: 10.1523/JNEUROSCI.4369-03-2004
Janik, V. M., & Slater, P. J. B. (1997). Vocal learning in mammals. En P. Slater, J. Rosenblatt, Ch. Snowdon, & M. Milinski (Eds.), Advances in the study of behavior. Vol. 24 (pp. 59–99). San Diego, CA: Academic Press.
Kiya, T., Itoh, Y., & Kubo, T. (2008). Expression analysis of the FoxP homologue in the brain of the honeybee, Apis mellifera. Insect Molecular Biology, 17, 53–60. doi: 10.1111/j.1365-2583.2008.00775.x
Knörnschild, M. (2014). Vocal production learning in bats. Current Opinion in Neurobiology, 28, 80–85. doi: 10.1016/j.conb.2014.06.014
Kuhl, P., & Meltzoff, A. (1982). The bimodal perception of speech in infancy. Science, 218, 1138–1141. doi: 10.1126/science.7146899
Lai, C. S. L., Fisher, S. E., Hurst, J. A., Vargha-Khadem, F., & Monaco, A. P. (2001). A forkhead-domain gene is mutated in a severe speech and language disorder. Nature, 413, 519–523. doi: 10.1038/35097076
Lawton, K. J., Wassmer, T. L., & Deitcher, D. L. (2014). Conserved role of Drosophila melanogaster FoxP in motor coordination and courtship song. Behavioural Brain Research, 268, 213–221. doi: 10.1016/j.bbr.2014/04.009
Li, S., Wang, Y., Zhang, Y., Lu, M. M., DeMayo, F. J., Dekker, J. D., Tucker, P. W., & Morrisey, E. E. (2012). Foxp1/4 control epithelial cell fate during lung development and regeneration through regulation of anterior gradient 2. Development, 139, 2500–2509. doi: 10.1242/dev.079699
Mendoza, E., Colomb, J., Rybak, J., Pflüger, H.-J., Zars, T., & Scharff, C. (2014). Drosophila FoxP mutants are deficient in operant self-learning. Plos One, 9(6), e100648. doi: 10.1371/journal.pone.0100648
Murugan, M., Harward, S., Scharff, C., & Mooney, R. (2013). Diminished FoxP2 levels affect dopaminergic modulation of corticostriatal signaling important to song variability. Neuron, 80(6), 1464–1476. doi: 10.1016/j.neuron.2013.1009.1021
Santos, M. E., Athanasiadis, A., Leitão, A. B., DuPasquier, L., & Sucena, É. (2011). Alternative splicing and gene duplication in the evolution of the FoxP gene subfamily. Molecular Biology and Evolution, 28, 237–247. doi: 10.1093/molbev/msq182
Shimeld, S. M., Degnan, B., & Luke, G. N. (2010). Evolutionary genomics of the Fox genes: Origin of gene families and the ancestry of gene clusters. Genomics, 95, 256–260. doi: 10.1016/j.ygeno.2009.08.002
Strausfeld, N. J., & Hirth, F. (2013). Deep homology of arthropod central complex and vertebrate basal ganglia. Science, 340, 157–161. doi: 10.1126/science.1231828
Vargha-Khadem, F., Gadian, D. G., Copp, A., & Mishkin, M. (2005). FOXP2 and the neuroanatomy of speech and language. Nature Reviews Neuroscience, 6, 131–138. doi: 10.1038/nrn1605
Watkins, K. E., Dronkers, N. F., & Vargha-Khadem, F. (2002). Behavioural analysis of an inherited speech and language disorder: Comparison with acquired aphasia. Brain, 125, 452–464. doi: 10.1093/brain/awf058
Wohlgemuth, S., Adam, I., & Scharff, C. (2014). FoxP2 in songbirds. Current Opinion in Neurobiology, 28, 86–93. doi: 10.1016/j.conb.2014.06.009
Zhang, G., Li, B., Li, C., Gilbert, M., Jarvis, E., Wang, J. & Consortium, T. A. G. (2014). Comparative genomic data of the Avian Phylogenomics Project. GigaScience, 3, 26. doi: 10.1186/2047-217X-3-26
ACKNOWLEDGEMENTS:
We are thankful to Arturo and Anna Zychlinsky for helpful editorial comments, to Kate Watkins for providing nonsense words used to assess speech performance and to Peter Schatton for lending his artistic talent to create Figures 1 and 7 and Ezequiel Mendoza to modify Figure 5. We apologize to the authors whose findings are not referenced due to space constraints. Research performed in the lab of the authors benefited from funds provided by the Excellence Cluster Neurocure and the SFB 665 «Developmental Disturbances of the Nervous System».