Communication about genetic editing
CRISPR, between optimism and false expectations

Communication is essential in all areas of society, but communication in science is inescapable. Communicating means sharing, showing, teaching, and transferring knowledge about discoveries, observations, and findings both to colleagues and to society in general. That is why good communication must always accompany good science. CRISPR genetic editing tools allow us to modify, at will, the genome of any living organism, including our own species. In this text I review the different relevant communicative events in the short but intense life of these «molecular scissors», so called for their ability to cut the DNA molecule effectively and with precision.
Keywords: CRISPR, expectations, interpretation, uncertainty, science communication.
«The assumption that the field of genetic editing started with CRISPR is a misrepresentation»
The origins
Reading the explosion of texts about CRISPR (clustered regularly interspaced short palindromic repeats) genetic editing tools published over the last four or five years, one might think that they did not exist before 2012 and that they appeared suddenly, as if by chance. However, such coincidences are rare in science.

In 2012, two research teams suggested that the same CRISPR system used by bacteria to defend themselves from viruses could be used to modify the DNA sequence of any organism. The Lithuanian researcher Virginijus Siksnys (below) collected his observations and submitted them to a journal. Jennifer Doudna (above) and Emmanuelle Charpentier (below) reached the same conclusion but managed to publish their paper in a different prestigious journal a couple of months before Siksnys. / © The Royal Society / Duncan Hull
If we condense the history of genetic editing tools into the last five years, an important communication problem is revealed. We can look for different origins, but genetic editing – that is, the precise and specific modification of genetic sequences at the will of researchers – started in 1985 with Oliver Smithies. He was the theorist of homologous recombination and an expert on how to exchange one, essentially identical, DNA sequence with another, and in so doing also removing any other sequence attached to the first; he saw that cutting the DNA sequence that he wanted to integrate into the corresponding homologous gene (in the genome sequence that shared the same letters as the donor molecule) very significantly increased the probability of correct insertion (Smithies, Gregg, Boggs, Koralewski, & Kucherlapati, 1985). Smithies was one of the three researchers who received the Nobel Prize in Medicine in 2007 for describing the method to inactivate specific genes in mice using pluripotent embryonic stem cells. These cells are erroneously called «mother» cells in Spanish, Catalan, and Galician, but the term has been successful in communication and even Spanish scientists use it regularly.
Years later French researchers from the Pasteur Institute used Smithies’ observation with the genome itself and discovered some enzymes called meganucleases, which can sever the genome at a precise spot in a unique way. Then, after 2001, zinc-finger nucleases (ZFNs) were discovered and these leapt onto the scene in the media in 2009 when they were first used to direct the specific inactivation of murine genes. Two years later, other enzymes, called transcription activator-like effector nucleases (TALENs), would be discovered in nature. These use pathogenic microorganisms from plants to revert the metabolism of the infected cells to their advantage. TALENs had their moment of glory until 2013, when the CRISPR tools were first used in genetic editing experiments with mammalian cells, specifically human cells (Fernández, Josa, & Montoliu, 2017). Thus, this is the story so far; therefore, the assumption that the field of genetic editing started with CRISPR is a misrepresentation.
Often, relatively few people are interested in the communication of scientific advances: generally, only those who work specifically in the field in which the findings are made. It is amazing to think how such relevant advances in genetic editing went virtually unnoticed by the general public for nearly 30 years.

Virginijus Siksnys / © Gie2016
CRISPR
What happened with CRISPRs? We learned about them in 2013, because of their spectacular and unexpected applications, which were both surprising and frightening, but they had been there for over 25 years. Why did communication about them fail? Why did they not receive the space and interest they deserved sooner? Again, this is a case of specialised basic knowledge, of discoveries we fail to read from a different angle until we were shown their surprising and life-changing applications.
In 1987, Japanese microbiologists discovered a repetitive DNA sequence while researching a chromosome fragment from Escherichia coli, bacteria which live in our intestines. They reported this strange finding but did not consider it important. Other Dutch microbiologists also found repetitive blocks of DNA sequences in different bacteria, Mycobacterium, responsible for tuberculosis and which, in evolutionary terms, are very different from E. Coli. They decided to use these repeats in the genome to classify different isolates from the bacteria with different numbers of repeats.

Emmanuelle Charpentier / © Bianca Fioretti, Hallbauer i Fioretti
At the beginning of the nineties, for his PhD work, Francisco Juan Martínez Mojica (Francis Mojica) carried out experiments with microorganisms called archaea, which live in extreme environments such as the Santa Pola salt marshes (Alicante, Spain). Archaea are prokaryotes (single cell microorganisms without a nucleus) like bacteria, but they are very different to them. Mojica also found repeats like those reported by Japanese and Dutch colleagues in Escherichia and Mycobacterium in archaea. He also realised that, if three such disparate microorganisms so evolutionarily separated from each other shared these repetitive sequences, they might be useful and fulfil some function so that evolution would preserve them. Therefore, he decided to devote the rest of his professional career to understanding them. This decision put CRISPR (a name coined by Mojica in 2001 for these sequence repeats) on the path to becoming one of the most transformative tools in biology. Meanwhile, the general public and researchers not associated with CRISPR were unaware that a new revolution was brewing and that it would explode twelve years later.
«Mojica noticed that the bacteria carrying fragments of the genome of certain viruses were resistant to infection by those viruses»
In 2003, Mojica had his great eureka moment. Reviewing unique segments between repeats in the DNA sequence (called «spacers» until then) he found similarities with the genome sequence of bacteriophages, viruses that infect bacteria. Even more importantly, he noticed that the bacteria carrying fragments of the genome of certain viruses were resistant (immune!) to infection by those viruses. In other words: he had just discovered an immune system in bacteria, an adaptive one that can learn and which was based on genetics, i.e., transmitted from bacteria to offspring bacteria. He confirmed this observation in other bacteria and then wrote and submitted a paper to the best scientific journals. However, they rejected his observations. Another serious communication problem and another great failure of the publication system. Three years went by with the paper bouncing from journal to journal, from rejection to rejection, until in 2005 he ended up publishing his data in a worthy but lesser journal, far from the spotlight and the most prominent covers (Mojica, Díez-Villaseñor, García-Martínez, & Soria, 2005). Trying to communicate his incredible results almost cost Mojica his career and, when they were published, they were again widely ignored by the scientific community and society. It was just another paper published in one of many journals.
Two years later, in 2007, French researchers carried out the experiment Mojica had been unable to: they transferred segments of sequences similar to virus genomes between different bacteria and in so, also transferred resistance to them. This experimentally verified Mojica’s initial observation. However, this publication would still not be enough to popularise the possibilities of genetic editing.

The communication of results, the manner and timing of their communication, the role of the narrator, and the communication channel used all have relevant consequences for the entire process. / © Sean Winters
In 2012 we had already accumulated a lot of knowledge about CRISPR’s systems because of their discovery in archaea by Mojica and the description of elements constituting a bacterial defence system by the French team more than twenty years before. Then, two research teams suggested that the same CRISPR system used by bacteria to defend themselves from viruses could be used to modify the DNA sequence of any organism at will, whether plant or animal – or even human. A Lithuanian researcher, Virginijus Siksnys, was probably the first to notice this new application of the elements constituting the CRISPR system. He collected his observations and sent them to a journal that would end up delaying their publication (Gasiunas, Barrangou, Horvath, & Siksnys, 2012).
Meanwhile, a collaboration between Jennifer Doudna and Emmanuelle Charpentier brought about by circumstance (they were working together to co-direct a postdoctoral researcher, but had worked independently until then and have not worked together again since) led them to the same conclusion as Siksnys. But, although they submitted after Siksnys they were able to use their privileged position to publish their paper in a prestigious journal in record time, just a couple months before Siksnys (Jinek et al., 2012). This communication strategy placed the two researchers in the hall of fame and condemned the Lithuanian microbiologist to ostracism. Very few now remember him: yet another major communication problem.

Thanks to articles published on blogs, open science and social networks have also been essential to uncovering incorrect information. / © Greg Emmerich
Naturally, the institution where the two researchers worked at the time decided to keep their industrial rights over the use of CRISPR genetic editing tools, made entirely in the laboratory with bacteria, before publishing their observations. Thus, Berkeley University registered a patent application. However, their paper published in Science in August 2012 did not prove that these CRISPR tools can be used to edit the genes of mammal cells – of human cells. It only proposed doing so.
Several months later, Feng Zhang entered the scene. He was a researcher at the Broad Institute at Boston MIT who, in January 2013, proved for the first time that bacteria CRISPR tools could indeed be used as genetic editors in human cells (Cong et al., 2013). His work coincided with the work of another researcher, George Church, also from Boston, who published his findings in the same issue of the journal Science (Mali et al., 2013). The Broad Institute, of course, also protected their researchers’ breakthrough and applied for the corresponding patent in late 2012, months after Doudna and Charpentier filed for theirs. At that time, an extraordinary event occurred which irreversibly marked the subsequent history of CRISPR. Broad chose to use a riskier fast patent-evaluation system that leaves no opportunity for corrections and has a significantly higher monetary cost. This coincided with a substantial change in the US Patent Office: they stopped granting patents to «the first to file an idea» and started giving them to «the first to prove its use». Doudna and Charpentier registered their patent before Zhang, but he was the first to prove that the CRISPR tools could be used for genetic editing in mammals.
The Patent Office granted the patent to the Broad Institute in 2014. Berkeley University filed a lawsuit for patent interference, because they believed that Zhang had copied the idea, or at least, his experiments to prove the use of CRISPR in human cells had been based on their published experiments. Zhang and the Broad Institute defended themselves indicating that they had followed a parallel path almost at the same time as Doudna and Charpentier’s. Finally, in early 2017, a US court rejected Berkeley’s interference lawsuit and confirmed that the Broad Institute was the owner of the patent1. This dispute became more complicated due to a review of the matter written by Eric Lader, director of the Broad Institute, who openly sided with Feng Zhang, his centre’s researcher, and blurred the role played by Doudna and Charpentier (Lander, 2016). The two researchers responded to his article in other media (Vence, 2016).
Once again, the communication of results, the manner and timing of their transmission, the role of the narrator, and the communication channels used had relevant consequences for the entire process.
«Doudna and Charpentier registered their patent before Zhang, but he was the first to prove that CRISPR tools could be used for genetic editing in mammals»
CRISPR communication
In 2013, the effectiveness of CRISPR in editing human or mouse cells was not only proven for the first time, these tools were also used in pioneering work to create mutant mice and zebrafish. However, all these breakthroughs and applications collected together many basic scientific findings made in the eighties by microbiologists, biochemists, and molecular biologists whose previous work had facilitated the spread of this new technology around the world (Mojica & Montoliu, 2016).
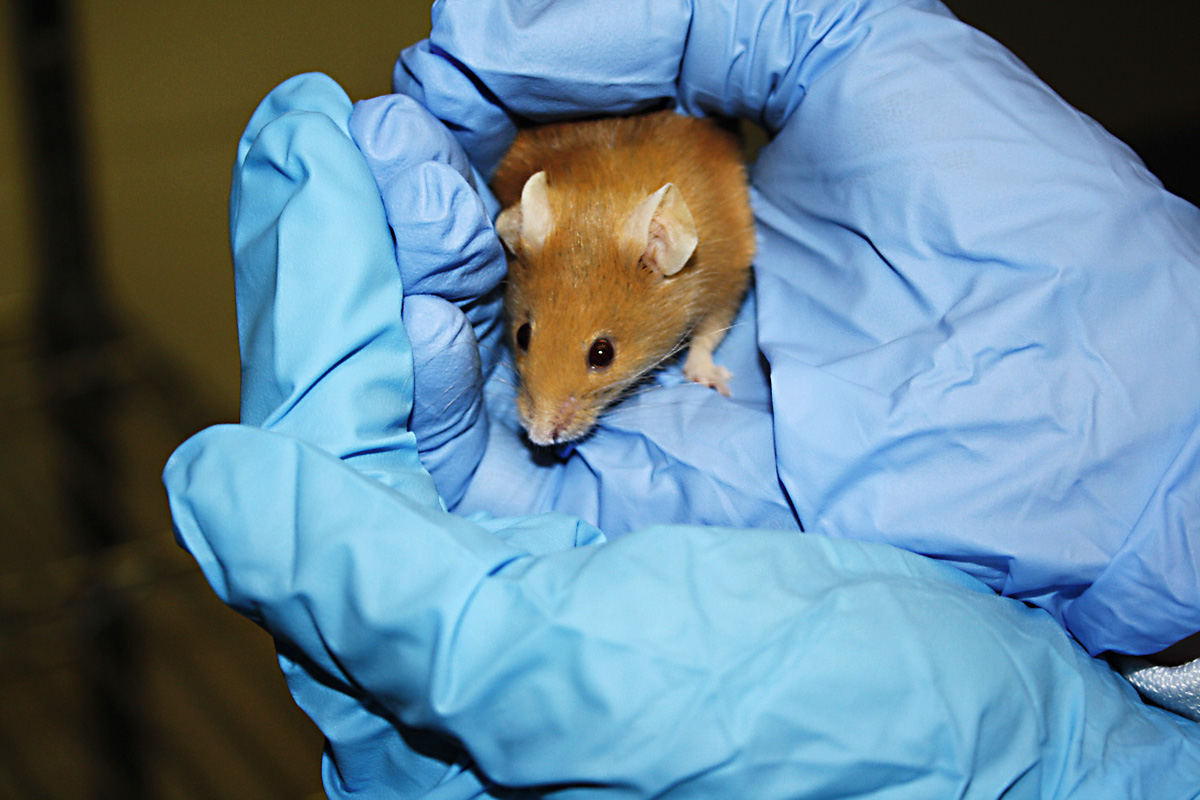
In the year 2013, the effectiveness of CRISPR in editing human or murine cells was not only proven for the first time, these tools were also used in pioneering work to create mutant mice and zebrafish. / © SforResearch
CRISPR tools have been used since 2013 in almost every field, from biology and biomedicine to biotechnology. At the beginning of 2016, three independent but related papers were published precisely in this field, proposing a novel administration method for CRISPR tools in model mice for a serious degenerative disease, Duchenne muscular dystrophy (Nelson et al., 2016). This work communicated, for the first time, that it was possible to restore the mutated gene causing a congenital disease in a significant number of cells of the affected organ – muscle fibres in this case – and this opened the possibility of using CRISPR as a somatic cell gene therapy tool to correct the mutation in affected cells. Undoubtedly the communication of this work has generated enormous expectations among the millions of patients around the world affected by a congenital disease. However, going from preclinical experiments with laboratory animals to clinical trials with patients is a step that requires extreme caution. We still cannot control the process of mutated gene correction well enough. The accuracy of CRISPR tools does not translate to the systems of cell repair that restore the DNA cut. These repair systems tend to generate a lot of genetic variability: many different types of DNA molecules among which the desired correct sequence can usually be found, albeit among many other sequences. This uncertainty, i.e., the genetic indeterminacy in correcting mutations in specific genes, has been confirmed in any genetic editing experiments in animals, whether in mice (Seruggia, Fernández, Cantero, Pelczar, & Montoliu, 2015) or human embryos (Liang et al., 2015).
© Concha Molina
Very recent experiments seem to suggest that mosaicism (the coexistence of two or more cell populations with a different genetic composition in the same individual) can be controlled in human embryos. However, the pioneering work describing the genetic correction of the mutation in a gene associated with a congenital cardiomyopathy by using CRISPR (Ma et al., 2017) has already been met with a response from several laboratories suggesting alternative, less optimistic, explanations for this apparent removal of genetic uncertainty. The genetic correction system itself might mean that the researchers were unable to analyse mutations in the same way they had initially done, making them believe that the gene mutation was corrected when in reality, they were simply unable to detect the mutated gene (Egli et al., 2017). Once again, the communication of potential advantages and solutions to a genetic problem might need to be revised, considering new results and new interpretations of previous results.
Additionally, CRISPR tools may cut DNA in sequences that are similar (rather than identical) to the ones that were initially planned, although this possibility has decreased as more specific guides using more powerful bioinformatics software have been designed. In fact, cuts at other points in the genome are very rare, or non-existent, in animal experiments (Seruggia et al., 2015). That is why another publication, in mid-2017 by American researchers, was so surprising. It described the existence of thousands of unexpected mutations in mice after a genetic editing experiment using CRISPR (Schaefer et al., 2017). This paper sparked great international controversy because it put a stop on all therapeutic hopes for CRISPR: who would want to use a therapeutic strategy associated with thousands of unexpected and uncontrolled mutations?

© Agencia Sinc
This new communication problem even affected the stock value of publicly traded companies that work with genetic editing, whose shares plummeted after such negative results. However, the findings were not true; just a few days later, evidence and alternative explanations started to pile up proving that the mice used by the authors were different to those initially used in the genetic editing experiment to analyse the DNA sequences (Iyer et al., 2018). Because of this difference, all the changes they manifested, and which this publication attributed to specificity problems derived from the use of CRISPR tools, had a much simpler explanation: the wrong genetic information had been used. It came from mice that were similar, but not identical, to the ones used in the initial experiment. In this case, social communication tools – social networks and blogs – were essential to quickly countering the apparently negative results reported in the paper with an alternative explanation. In a few weeks, we went from initial shock to a degree of embarrassment for the researchers involved, whose rookie mistake of comparing two different mouse lineages could have completely deposed the CRISPR tools from their privileged position in the treatment of genetic diseases – i.e., for gene therapy.
Social networks and open science (thanks to articles published on blogs) were also essential to uncovering incorrect information in results published in May 2016 by a Chinese research team, who had announced that they had discovered new genetic editing tools – apparently much better than CRISPR. But the new tool, called NgAgo, seemed to work only in the laboratory that described it. In just two months, the first reviews appeared on different blogs, and soon after, researchers from around the world verified that the published results were not reproducible (Khin, Lowe, Jensen, & Burgio, 2017). Finally, in August 2017, the journal responded to the general outcry by retracting the publication.

© Concha Molina
China was not only burdened by frauds like NgAgo, it was also responsible for pioneering experiments using CRISPR in human embryos (Liang et al., 2015), which explored the limits of the technique for correcting certain mutations. They noted – as in the rest of species tested – the uncertainty of results and phenomena such as genetic mosaicism and the possibility of altering other genome sequences, similar to those of the targets. Moreover, initially through press releases with partially published results, we have also learned about China’s world leadership in the race to use CRISPR tools to treat patients, by editing the genes of lymphocytes taken from the blood of cancer patients (Su et al., 2016). However, the first patient treated in vivo with genetic editing tools (although with ZFN, not CRISPR) was a Californian man, as part of a clinical trial approved in the USA and whose results were released to the public in 2017.
In this paper I have related how communication topics have strongly impacted the development, knowledge, and application of the new CRISPR genetic editing tools. Similarly, I have also discussed the most controversial communicative aspects, especially the communication of apparently discouraging results, which have appeared on several occasions in the short history of genetic editing. But, similarly, this has favoured the publication of thousands of immediate responses by other researchers, who, using every communication channel within their reach, have managed to counter the pessimism or false hopes from such results until they can be properly analysed and adequately understood by other researchers and by society in general.
1. On 10 September 2018, the US Court of Appeals for the Federal Circuit finally awarded the pivotal CRISPR patent to the Broad Institute. (Go back)
References
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., … Zhang, F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science, 339(6121), 819–823. doi: 10.1126/science.1231143
Egli, D., Zuccaro, M., Kosicki, M., Church, G., Bradley, A., & Jasin, M. (2017). Inter-homologue repair in fertilized human eggs? BioRxiv, 28 August 2017. doi: 10.1101/181255
Fernández, A., Josa, S., & Montoliu, L. (2017). A history of genome editing in mammals. Mammalian Genome, 28(7–8), 237–246. doi: 10.1007/s00335-017-9699-2
Gasiunas, G., Barrangou, R., Horvath, P., & Siksnys, V. (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the USA, 109(39), E2579–E2586. doi: 10.1073/pnas.1208507109
Iyer, V., Boroviak, K., Thomas, M., Doe, B., Ryder, E., & Adams, D. (2018). No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. BioRxiv, 9 February 2018. doi: 10.1101/263129
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096), 816–821. doi: 10.1126/science.1225829
Khin, N. C., Lowe, J. L., Jensen, L. M., & Burgio, G. (2017). No evidence for genome editing in mouse zygotes and HEK293T human cell line using the DNA-guided Natronobacterium gregoryi Argonaute (NgAgo). PLOS One, 12(6), e0178768. doi: 10.1371/journal.pone.0178768
Lander, E. S. (2016). The heroes of CRISPR. Cell, 164(1–2), 18–28. doi: 10.1016/j.cell.2015.12.041
Liang, P., Xu, Y., Zhang, X., Ding, C., Huang, R., Zhang, Z., … Huang, J. (2015). CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell, 6(5), 363–372. doi: 10.1007/s13238-015-0153-5
Ma, H., Marti-Gutierrez, N., Park, S.-W., Wu, J., Lee, Y., Suzuki, K., … Mitalipov, S. (2017). Correction of a pathogenic gene mutation in human embryos. Nature, 548(7668), 413–419. doi: 10.1038/nature23305
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., … Church, G. M. (2013). RNA-guided human genome engineering via Cas9. Science, 339(6121), 823–826. doi: 10.1126/science.1232033
Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J., & Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of Molecular Evolution, 60(2), 174–182. doi: 10.1007/s00239-004-0046-3
Mojica, F. J., & Montoliu, L. (2016). On the origin of CRISPR-Cas technology: From prokaryotes to mammals. Trends in Microbiology, 24(10), 811–820. doi: 10.1016/j.tim.2016.06.005
Nelson, C. E., Hakim, C. H., Ousterout, D. G., Thakore, P. I., Moreb, E. A., Castellanos-Rivera, R. M., … Gersbach, C. A. (2016). In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science, 351(6271), 403–407. doi: 10.1126/science.aad5143
Schaefer, K. A., Wu, W. H., Colgan, D. F., Tsang, S. H., Bassuk, A. G., & Mahajan, V. B. (2017). Unexpected mutations after CRISPR-Cas9 editing in vivo. Nature Methods, 14(6), 547–548. doi: 10.1038/nmeth.4293 (Retracted in 2018, Nature Methods, 15, 394. doi: 10.1038/nmeth0518-394a).
Seruggia, D., Fernández, A., Cantero, M., Pelczar, P., & Montoliu, L. (2015). Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR-Cas9-mediated mutagenesis. Nucleic Acids Research, 43(10), 4855–4867. doi: 10.1093/nar/gkv375
Smithies, O., Gregg, R. G., Boggs, S. S., Koralewski, M. A., & Kucherlapati, R. S. (1985). Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature, 317(6034), 230–234. doi: 10.1038/317230a0
Su, S., Hu, B., Shao, J., Shen, B., Du, J., Du, Y., …, Liu, B. (2016). CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Scientific Reports, 6, 20070. doi: 10.1038/srep20070
Vence, T. (2016, 19 January). “Heroes of CRISPR” disputed. The Scientist. Retrieved from https://www.the-scientist.com/?articles.view/articleNo/45119/title/-Heroes-of-CRISPR--Disputed