Building brains that can evolve
Challenges and prospects for evo-devo neurobiology
Evo-devo biology involves cross-species comparisons of entire developmental trajectories, not just of adult forms. This approach has proven very successful in general morphology, but its application to neurobiological problems is still relatively new. To date, the most successful area of evo-devo neurobiology has been the use of comparative developmental data to clarify adult homologies. The most exciting future prospect is the use of comparative developmental data to understand the formation of species differences in adult structure and function. An interesting «model system» for this kind of research is the quest to understand why the neocortex folds in some species but not others.
Keywords: brain, evolution, development, homology, cortical folding.
I was taught that the trick to doing good science is to find a set of questions that are both interesting and solvable, given the methods currently available (Medawar, 1967). I also learned that this set of questions tends to change over time, as old questions are answered or lose their gloss, and novel methods make new questions addressable. Given these lessons, I periodically take stock of where my field of inquiry has been and where it is going. This essay reflects those thoughts, though it is written from a larger perspective, surveying the current state and prospects of evo-devo neurobiology, which combines the study of brain evolution with that of brain development.
Evo-devo in general biology
For much of the twentieth century, evolutionary biology was dominated by a view of evolution that essentially ignored development and treated evolutionary change as if ancestral adults transformed directly into adult descendants. For example, in widely publicized diagrams of horse evolution, small ancestral adults were shown as transforming into much larger adult forms. The fact that every adult horse develops from an embryo and leaves offspring that has to grow before it reaches adult size was deemed largely irrelevant to the evolutionary storyline. As the century progressed, evolutionary biologists did become interested in the mechanism underlying evolutionary changes in adult morphology, but they focused their analyses almost exclusively on changes in gene frequencies, largely ignoring the role of genes in organismal development. As Theodosius Dobzhansky, a founding father of the modern synthesis in evolutionary biology, put it in 1951: «The study of mechanisms of evolution falls within the province of population genetics» (cited in Gilbert, Opitz, & Raff, 1996). Thus, the modern synthesis combined population genetics with classical comparative morphology but had little or no use for embryology.
«The modern synthesis combined population genetics with classical comparative morphology but had little or no use for embryology»
One likely reason for this omission was that Ernst Haeckel and his followers in the late 1800s had fatally oversimplified the link between development (ontogeny) and evolutionary change (phylogeny). They had argued that individual organisms in their development pass through the adult stages of their ancestors or, more succinctly, that «ontogeny recapitulates phylogeny» (see Gould, 1977). According to this view, mammalian embryos go through a stage of development at which they have gills that correspond to the gills of ancient fishes and amphibians from which mammals evolved. In Haeckel’s view, adult mammals do not retain these gills simply because they continue to develop until the gills have been transformed into other structures (e.g., jaw and middle ear bones). In essence, Haeckel and his followers imagined evolutionary change to occur solely by the addition of new stages to the end of an ancestral ontogeny. If this is true, then each species must, in its embryological development, pass through the adult stages of its ancestors in chronological order.
However, Haeckel’s view was far too limited. A mountain of comparative embryological data has now revealed that evolutionary transformations sometimes involve the deletion of ancestral adult stages (sometimes called paedomorphosis) or, quite frequently, changes in the direction of development «midstream» (i.e., before the ancestral adult stage was reached). The latter view, of development as diverging between species, dates back to Karel Ernst von Baer in the middle of the nineteenth century and thus predates Haeckel’s main work, but it was slow to catch on, perhaps because Von Baer had steadfastly opposed Darwin’s theory of evolution. In any case, whenever the developmental trajectory of a descendant species ends prematurely or diverges from that of its ancestor, individual development fails to recapitulate phylogeny (Figure 1). Instead of ontogeny recapitulating phylogeny, the most common pattern is that different species resemble each other rather closely during some early stage of embryonic development (often called the phylotypic stage) and then diverge as they grow up. Thus, embryonic mammals do not really have gills; instead they have «pharyngeal slits» that closely resemble those of other embryonic vertebrates. As these pharyngeal slits (and the bars between them) develop, they assume very different forms in different vertebrate lineages.
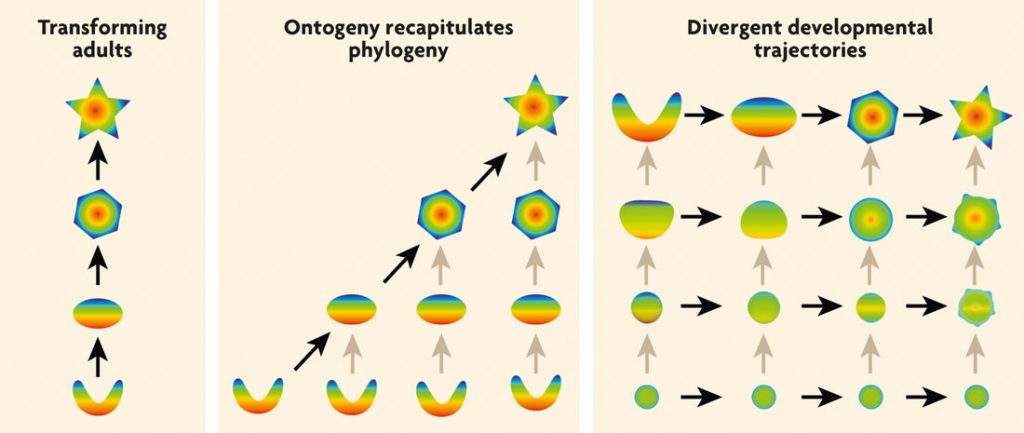
Figure 1. Changing views on the relationship between development and evolutionary change. Most people consider evolutionary change to consist of ancestral adults transforming directly into adult descendants (left). Ernst Haeckel and his followers tried to integrate evolution with development by claiming that «ontogeny recapitulates phylogeny», meaning that successive stages of an individual’s development correspond to the adult stages of successive ancestors (middle). In the more modern evo-devo perspective, evolutionary change may involve changes at any step along an ancestral ontogeny, with developmental trajectories frequently diverging between species. As a result, early stages of development tend to be more similar across species than later stages. Black arrows indicate transformations between adult forms or similar stages; gray arrows indicate developmental transformations. / Georg F. Striedter
A second problem with Haeckel’s view was that he saw development as the mechanism that drove evolutionary change. This view makes sense in light of Haeckel’s belief that changes in an individual’s development can be passed on to its offspring (i.e., the inheritance of acquired characteristics), but Haeckel’s view was difficult to integrate into the modern synthesis of evolutionary biology, because in this synthesis, developmental differences were viewed as the outcome of evolution, not as its cause. As mentioned earlier, the creators of the modern synthesis considered changes in gene frequencies to be the mechanistic drivers of evolutionary change; the causal links between genes and development remained obscure and neglected. This situation changed toward the end of the twentieth century, when developmental biology became increasingly molecular, exploring and manipulating gene expression in embryos. Once this work became comparative, extending beyond an elite group of «model species», it became possible to study the molecular mechanisms underlying evolutionary divergence in development. Through this work, developmental biology gradually became integrated with evolutionary biology, complementing population genetics, and evo-devo biology was born (Gilbert et al., 1996). This fledgling field then expanded rapidly and is thriving today, as evidenced by dedicated journals and grant review panels.
I, too, became enthralled with the evo-devo approach in the 1980s. I liked the idea of getting at the molecular mechanisms of evolutionary change by comparing the ontogenies of diverse species, identifying specific evolutionary changes in developmental trajectories, correlating those morphological changes with changes in gene expression and, ultimately, using experimental manipulations to «re-create» those evolutionary changes in the laboratory. I was impressed, for example, by experimental manipulations that induced chicken embryos to grow some teeth (Kollar & Fisher, 1980), even though living birds lost all their adult teeth millions of years ago. More recently, I marveled at research that compared beak development across diverse Galápagos finches and then tested a hypothesis about the molecular mechanisms underlying this variation by manipulating beak development in embryonic chicks (Abzhanov, Protas, Grant, Grant, & Tabin, 2004). Numerous studies of this kind have now proven that the evo-devo approach can be fruitfully applied to the evolution of beaks, limbs, fins, and bones. But would it work for brains? Would the brain’s immense complexity stifle evo-devo neurobiologists? To some extent it did, at least initially. However, evo-devo neurobiology has grown substantially in recent years and promises good future growth.
Evo-devo neurobiology
To date, most evo-devo neurobiologists have focused their research not on explaining species differences in brains, but on finding unsuspected, cryptic similarities. Very successful in this endeavor have been Luis Puelles and his colleagues (e.g., Puelles et al., 2000). They compared the expression patterns of numerous genes in the brains of embryonic mammals and birds, as well as a few other vertebrate groups, and discovered that those patterns are remarkably similar across these species, even though the adult brains are very different, especially in the forebrain. These findings are consistent with the general pattern of developmental divergence (rather than recapitulation) that I discussed earlier. Moreover, the similarities in embryonic gene expression allowed Puelles et al. (2000) to infer numerous homologies – corresponding features that can be traced back to a single evolutionary origin – that had remained unclear as long as only adult forms were considered. The same general approach has also been used to homologize major brain regions across the vertebrate-invertebrate divide (Denes et al., 2007; Holland, Holland, Williams, & Holland, 1992).
Although similarities in embryonic gene expression have clarified a few homologies, many remain controversial. One reason for these seemingly endless debates is that not all genes are equally good as «markers of homology». Most useful are genes that expressed only in the structures of interest and known to be involved in controlling their development and adult fate (Medina, Abellán, & Desfilis, 2013). When such ideal «markers» are not available, the best approach is to analyze the expression of many different genes – ideally, the entire transcriptome – and then compare the species statistically. Different statistics may yield different answers, but the statistical approach is certainly preferable to focusing on just the genes that support or contradict a particular hypothesis (i.e., cherry-picking). Such transcriptome comparisons have already been applied to adult brain regions, but significant advances will likely come when these analyses are extended to embryonic brain regions. After all, as noted earlier, embryonic brains are generally more similar across species than adult brains. Furthermore, genes expressed during embryonic development are more likely to participate in the regulatory gene networks that are thought to specify «character identity».
«Numerous studies have now proven that the evo-devo approach can be fruitfully applied to the evolution of beaks, limbs, fins, and bones. But would it work for brains?»
Once brain region homologies have been identified, it becomes possible to correlate differences in the level and spatial extent of embryonic gene expression with adult species differences in brain morphology. For example, work in my own laboratory has shown that adult parrots have an unusually small midbrain, relative to other birds, and that this adult species difference correlates with a species difference in the expression pattern of a gene that is involved in midbrain development (McGowan, Kuo, Martin, Monuki, & Striedter, 2011). Similarly, other scientists have shown that, among a group of teleost fishes, adult differences in the size of specific forebrain areas correlate with species differences in the expression of genes involved in forebrain patterning. Indeed, manipulating the expression of these genes can re-create (i.e., phenocopy) some of the adult species differences (Sylvester et al., 2010). What remains unknown in these studies is the molecular cause of the species differences in embryonic gene expression. How did evolution alter the regulation of these genes? Were some genes or regulatory DNA sequences added to (or deleted from) ancient gene regulatory networks? Answering such questions is often difficult because gene regulatory elements may be located quite far from the regulated gene. Moreover, some differences in gene expression may result from changes to DNA that do not involve changes in the DNA sequence (i.e., epigenetic modifications) but can nonetheless be transmitted across generations.
Such challenges are bound to hinder progress in evo-devo neurobiology, but it would be a mistake to argue that genomic and epigenomic explanations are the only acceptable kind of mechanistic explanation for evolutionary change in brain morphology. After all, brain development can be studied at several different levels of analysis – ranging from gene regulation to tissue formation – and each level can be considered a «causal mechanism» in its own right. For example, my own research has shown that the expansion of the telencephalon in parrots and songbirds, relative to other birds, is caused by an evolutionary delay in neurogenesis (which increases the number of telencephalic precursor cells; Charvet & Striedter, 2009). We do not know the molecular mechanism underlying this change in neurogenesis timing, but our finding is nonetheless mechanistic. Biology is hierarchically organized, and different scientists prefer to study different levels; that is why one scientist’s mechanism tends to be another scientist’s phenomenon that needs to be explained. In the long run, science is greedy, wanting all questions answered, but at any given moment, only some answers are actually attainable. As summarized in the next section, an excellent model system for illustrating this multi-tiered nature of evo-devo neurobiology is the folding of the neocortex in a subset of mammalian brains.
Cortical folding: A model system for evo-devo neurobiology
The largest part of the cerebral cortex, the neocortex, exhibits complex folds in humans and in other mammals with large brains. In fact, almost all mammalian brains larger than about 10 g tend to have a folded neocortex, whereas smaller brains do not. Moreover, the degree of cortical folding increases predictably with cortical surface area (especially when cortical surface area is multiplied by the square root of cortical thickness). Overall, these data indicate that the evolution of cortical folding is causally linked to evolutionary increases in neocortex area. Put differently, neocortical folding evolved whenever mammals increased the extent of their neocortex beyond a certain limit.
The functional significance of cortical folding remains debatable. It is clear, however, that without the folds, the neocortex would form a huge fluid-filled balloon that would complicate childbirth and be difficult to balance on the neck. In addition, cortical folding allows the axons that connect different parts of the neocortex to one another and the rest of the brain to be much shorter than they would be if the neocortex were balloon-shaped and smooth. Minimizing axon length, in turn, saves metabolic energy and increases the speed with which neurons in different brain regions can communicate with one another. Because of these functional benefits, cortical folding is surely adaptive, meaning that its evolution would have increased an individual’s odds of survival and reproduction. This «adaptationist» explanation for the evolution of cortical folding is not developmental in nature (i.e., not evo-devo), but it is nonetheless mechanistic; without folding, large neocortices could almost certainly not have evolved (unless they abandoned their sheet-like, cortical organization).
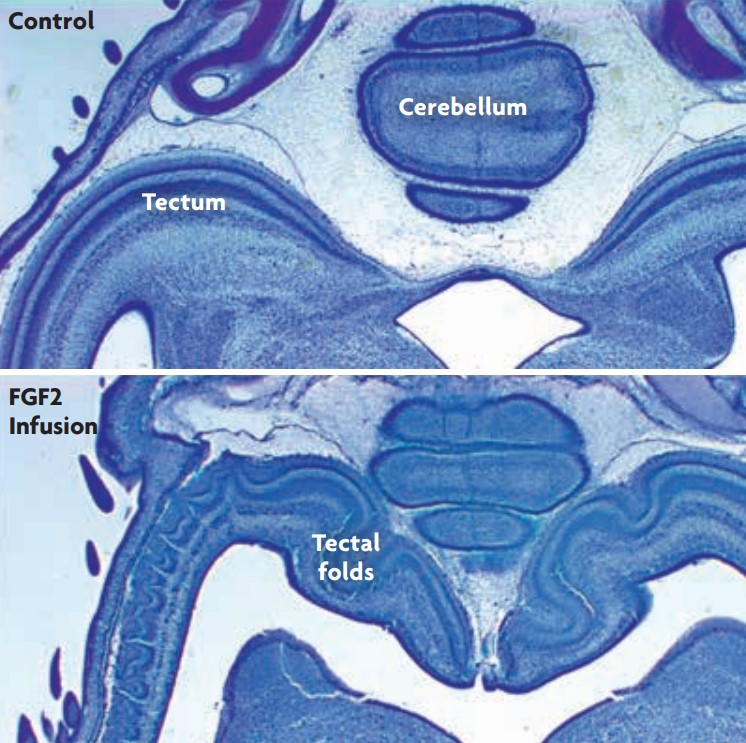
Figure 2. Experimentally induced folding of the optic tectum in birds. Infusing the transcription factor FGF2 into the cerebral ventricles of embryonic chickens delays tectal neurogenesis and, thereby, causes the optic tectum to become enlarged in volume and, especially, in surface area (McGowan et al., 2011). Remarkably, the enlarged optic tecta develop numerous folds, which normal tecta never do. We have proposed that these induced folds are a direct result of the tectum’s increased tangential expansion, which generates mechanical buckling forces. / Georg F. Striedter
From an evo-devo perspective, cortical folding is a developmental phenomenon that requires some sort of developmental explanation: why does the neocortex, which is smooth during early stages of development, begin to fold in those species in which the adult neocortex is folded? What forces generate the folds? We started thinking earnestly about this question in my laboratory after we manipulated chicken embryos into developing an enlarged optic tectum and, to our surprise, discovered that this enlarged tectum developed several folds (Figure 2; McGowan et al., 2012). Although we were working on birds rather than mammals, and focused on the tectum rather than the neocortex, we started to think that there might be some general principles that could explain tissue folding in diverse structures and species.
Guided by this idea and an extensive prior literature on cortical folding, we proposed that cortical folding in mammals results mainly from buckling forces that are generated when the cellular layer of the embryonic neocortex, called the cortical plate, expands in surface area (i.e., tangentially) more rapidly than the underlying tissue (Striedter, Srinivasan, & Monuki, 2015). As shown in modeling studies, such differential expansion leads to folding once the expansion has proceeded beyond a buckling threshold (which varies with the thickness and stiffness of the various layers). Additional mechanisms, such as the tension created by growing axons, may influence the specific locations where the folds form. Given this causal framework, the reason why the cortex folds more in some species than in others is that the cortical plate grows much larger (in area, not in thickness) in the species with highly folded cortices (Figure 3). The more it grows, relative to the underlying tissue, the greater the buckling forces, and the greater the degree of cortical folding.
«Most evo-devo neurobiologists have focused their research not on explaining species differences in brains, but on finding unsuspected, cryptic similarities»
What, then, causes species differences in the degree of cortical plate expansion? To answer this question, consider that the cortical plate is generated from progenitor cells that are located below the cortical plate (red zone in Figure 3). In species with small, smooth cortices, the progenitors initially divide to form additional progenitors, but soon begin to generate daughter cells that will not divide again; these post-proliferative cells then migrate radially into the emerging cortical plate, and differentiate into neurons. In contrast, in species with large and highly folded cortices, the cortical progenitors go through several additional rounds of cell division before they begin to generate post-proliferative cells. Because of these additional cell divisions, many more cells end up migrating into the cortical plate and forming the neocortex. Moreover, as young neurons migrate into the cortical plate, they push neighboring cells aside in such a way that the cortical plate spreads tangentially (i.e., expands in surface area) but does not thicken appreciably. On account of this «radial intercalation», evolutionary increases in the number of neocortical neurons are associated with major increases in cortical surface area (but only minor increases in cortical thickness). Those increases in cortical surface area, coupled with much smaller expansions of the underlying tissue, increase the buckling forces that generate cortical folds.
What evolutionary changes in the DNA cause cortical progenitors to divide more often in some species than in others? This question is unlikely to have a simple answer, because many different molecules are involved in cell cycle control, and it is certainly possible that the evolutionary expansion of the cortical progenitor pool is based on different molecular mechanisms in different lineages. However, the problem has been studied productively in one very interesting lineage, namely ours (Florio et al., 2015). This work has shown that the gene ARHGAP11B originated in the human lineage, shortly after it diverged from chimpanzees, through a partial duplication of an ancestral gene. Artificially expressing this gene in cortical progenitors of mice caused those progenitors to generate an unusually large number of «basal progenitors», thereby increasing the total size of the cortical progenitor pool. Most intriguingly, half of the mice expressing ARHGAP11B in their cortical progenitors ended up developing cortical folds that look quite similar to folds in human cortices. Thus, the de novo evolution of ARHGAP11B is likely to be a major molecular cause of cortical expansion and folding in the human lineage.
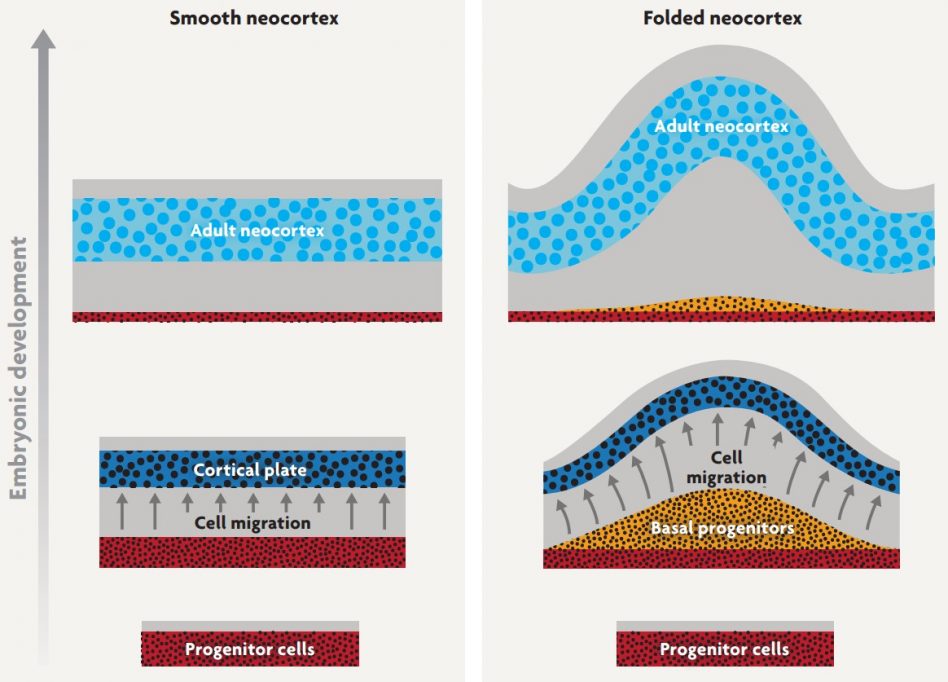
Figure 3. A developmental mechanism for the evolution of neocortical folds. When cortical progenitor cells stop dividing, they migrate radially away from the progenitor zone (faint red) to form a cell-dense cortical plate (dark blue). As development proceeds, the cortical plate expands and, eventually, gives rise to the adult neocortex (light blue). In species with folded neocortices (right), the progenitors divide more frequently, typically forming an extra layer of «basal progenitors». This expansion of the progenitor pool causes more neurons to migrate into the cortical plate. These migrating neurons insert themselves into the cortical plate in such a way that the plate expands tangentially much more than it thickens. Because the underlying tissue does not expand tangentially as fast, the cortical plate begins to buckle and fold. As the cortical plate matures into adult neocortex, the early folds become more prominent (Striedter et al., 2015) / Georg F. Striedter
Frontiers of evo-devo Neurobiology
As research on cortical folding and other evo-devo «model systems» progresses, the number of mechanisms underlying each phenomenon will probably increase. Some of these mechanisms will occupy different levels of analysis, making it important to be clear about how they relate to one another (Striedter, 1998). Even within a given level, multiple mechanisms will likely be revealed. For example, the evolution of ARHGAP11B is probably just one of many molecular mechanisms that lead to cortical expansion and folding, even within the human lineage. As this diversity of mechanisms comes into focus, it will be important to distinguish major causative factors – what some have called «mediators» – from minor, permissive ones – which one might call «modulators». Making this distinction will not always be easy, especially if different mechanisms predominate in different lineages, yet generate similar morphologies. Even if a well-defined experimental manipulation, such as the expression of ARHGAP11B in mice, produces a phenotype that resembles an evolutionary variant, this does not prove that the manipulated mechanism is the main cause of the natural phylogenetic change. One has to keep in mind that in biology there often is more than one way to solve a problem, more than one way to construct a given phenotype.
Still, the use of experimental manipulations to phenocopy at least some aspects of evolutionary change can be very useful. Particularly interesting would be to explore how the rest of the biological system responds to the specific manipulation. For example, I proposed some time ago that evolutionary increases in the size of one brain region, relative to the rest of the brain, might cause the enlarged area to project to a greater variety of targets and receive a greater variety of inputs (Striedter, 2005). This hypothesis is supported by some correlative data, but it has not yet been tested experimentally. To perform such a test one could, for example, express ARHGAP11B in the cortical progenitors of mice and then examine whether the enlarged (and folded) adult cortices make more axonal connections than they do in normal, wild-type mice. Support for the hypothesis would be exciting, because it would imply the existence of developmental mechanisms that help to integrate evolutionary changes in one part of the nervous system with the rest of the system, obviating the need for additional genomic changes to accomplish the integration.
«It would be a mistake
to argue that genomic and epigenomic explanations are the only acceptable kind of mechanistic explanation for evolutionary change in brain morphology»
More generally, it will be exciting to discover the extent to which experimental manipulations that specifically target one brain area or mechanism have cascading «downstream» effects on other brain regions and mechanisms, and to what extent these cascade effects are adaptive. I am fascinated, for example, by the finding that goats that are born without front legs will learn to walk bipedally and then develop adaptive changes in many parts of their body to accommodate this new form of locomotion (West-Eberhard, 2005). I suspect that nervous systems are also capable of such useful plasticity; indeed, this kind of developmental plasticity would have been adaptive in the sense that it promotes evolvability.
Just as interesting is the idea that developing nervous systems might be robust to many developmental perturbations, such as those produced by natural genetic or environmental variation within a population. The idea of such developmental «buffering mechanisms» dates back at least to Conrad Waddington (see Striedter, 1998) but the phenomenon is difficult to study because, after all, the relevant manipulations cause no adult phenotype. Still, the mechanisms underlying developmental robustness can be studied experimentally, and they are increasingly amenable to mathematical analysis and modeling (e.g., Lander, 2011). Overall, I expect that nervous systems have evolved to balance the need for developmental robustness with the need for evolvability. Understanding how this balance is achieved and managed over evolutionary time would, in my view, be a worthy ambition for evo-devo neurobiology.
REFERENCES
Abzhanov, A., Protas, M., Grant, B. G., Grant, P. R., & Tabin, C. J. (2004). BMP4 and morphological variation of beaks in Darwin’s finches. Science, 305, 1462–1465. doi: 10.1126/science.1098095
Charvet, C. J., & Striedter, G. F. (2009). Developmental origins of mosaic brain evolution: Morphometric analysis of the developing zebra finch brain. The Journal of Comparative Neurology, 514(2), 203–213. doi: 10.1002/ cne.22005
Denes, A. S., Jékely, G., Steinmetz, P. R. H., Raible, F., Snyman, H., Prud’homme, B., … Arendt, D. (2007). Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell, 129(2), 277–288. doi: 10.1016/j.cell.2007.02.040
Florio, M., Albert, M., Taverna, E., Namba, T., Brandl, H., Lewitus, E., … Huttner, W. B. (2015). Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science, 347, 1465–1470. doi: 10.1126/science.aaa1975
Gilbert, S. F., Opitz, J. M., & Raff, R. A. (1996). Resynthesizing evolutionary and developmental biology. Developmental Biology, 173(2), 357–372. doi: 10.1006/dbio.1996.0032
Gould, S. J. (1977). Ontogeny and phylogeny. Cambridge, MA: Harvard University Press.
Holland, P. W. H., Holland, L. Z., Williams, N. A., & Holland, N. D. (1992). An amphioxus homeobox gene: Sequence conservation, spatial expression during development and insights into vertebrate evolution. Development, 116, 653–661.
Kollar, E. J., & Fisher, C. (1980). Tooth induction in chick epithelium: Expression of quiescent genes for enamel synthesis. Science, 207, 993– 995. doi: 10.1126/science.7352302
Lander, A. D. (2011). Pattern, growth, and control. Cell, 144, 955–969. doi: 10.1016/j. cell.2011.03.009
McGowan, L., Kuo, E., Martin, A., Monuki, E. S., & Striedter, G. (2011). Species differences in early patterning of the avian brain. Evolution, 65, 907–911. doi: 10.1111/j.1558-5646.2010.01126x
McGowan, L. D., Alaama, R. A., Freise, A. C., Huang, J. C., Charvet, C. J., & Striedter, G. F. (2012). Expansion, folding, and abnormal lamination of the chick optic tectum after intraventricular injections of FGF2. PNAS, 109(S1), 10640–10646. doi: 10.1073/pnas.1201875109
Medawar, P. B. (1967). The art of the soluble. Londres: Methuen.
Medina, L., Abellán, A., & Desfilis, E. (2013). A never-ending search for the evolutionary origin of the neocortex: Rethinking the homology concept. Brain, Behavior and Evolution, 81(3), 150–153. doi: 10.1159/000348282
Puelles, L., Kuwana, E., Puelles, E., Bulfone, A., Shimamura, K., Keleher, J., … Rubenstein, J. L. R. (2000). Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx- 2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. The Journal of Comparative Neurology, 424, 409–438. doi: 10.1002/1096-9861(20000828)424:3<409::AID-CNE3>3.0.CO;2-7
Striedter, G. F. (1998). Stepping into the same river twice: Homologues as recurring attractors in epigenetic landscapes. Brain, Behavior and Evolution, 52, 218–231. doi: 10.1159/000006565
Striedter, G. F. (2005). Principles of brain evolution. Sunderland, MA: Sinauer Associates.
Striedter, G. F., Srinivasan, S., & Monuki, E. S. (2015). Cortical folding: When, where, how, and why? Annuals Reviews of Neuroscience, 38, 291–307. doi: 10.1146/annurev-neuro-071714-034128
Sylvester, J. B., Rich, C. A., Loh, Y.-H. E., von Staaden, M. J., Fraser, G. J., & Streelman, J. T. (2010). Brain diversity evolves via differences in patterning. PNAS, 107, 9718–9723. doi: 10.1073/pnas.1000395107
West-Eberhard M. J. (2005). Phenotypic accommodation: Adaptive innovation due to developmental plasticity. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 304, 610–618. doi: 10.1002/ jez.b.21071