When biology became engineering
Adopting standards for living systems
For decades, molecular biologists have been removing or inserting genes into all kinds of organisms with biotechnological intent or simply to generate fundamental knowledge. Synthetic biology (SynBio) goes one step further by incorporating conceptual frameworks from computing, electronics, and industrial design. This change makes it possible to conceive the creation of complex biological objects that were previously considered too difficult to assemble. To do this, the stages of any industrial production process must be adopted: design, construction of the components, assembly, and final manufacture. This objective requires standardisation of the physical and functional formats of the components involved, DNA assembly methods, activity measurements, and descriptive languages.
Keywords: synthetic biology, standards, repressilator, repository, orthogonality.
The founding effect: biology as seen by engineers
Although the history of synthetic biology (SynBio) as we know it goes back a long way, the birth of its contemporary version can be clearly traced back to the Massachusetts Institute of Technology (MIT) in the early 2000s. At the time, Tom Knight, a professor of artificial intelligence at MIT’s Department of Computer Science, started to formalise the idea of approaching biological systems with all the conceptual ammunition of electrical and industrial engineering, and using computer engineering as an interpretative framework. To do this, the fundamental abstractions that engineers use when analysing and designing objects of varying degrees of complexity had to be adapted to living entities. In this way, what we consider the central dogma of molecular biology is replaced by a highly abstract interpretative framework in which biological parts lead to devices that, in turn, lead to modules and systems (Figure 1). Thus, the evolutionary context as an explanation of the origin of biological functions is set aside and instead, all our effort is focused on the relational logic that makes living systems work here and now (De Lorenzo, 2018).
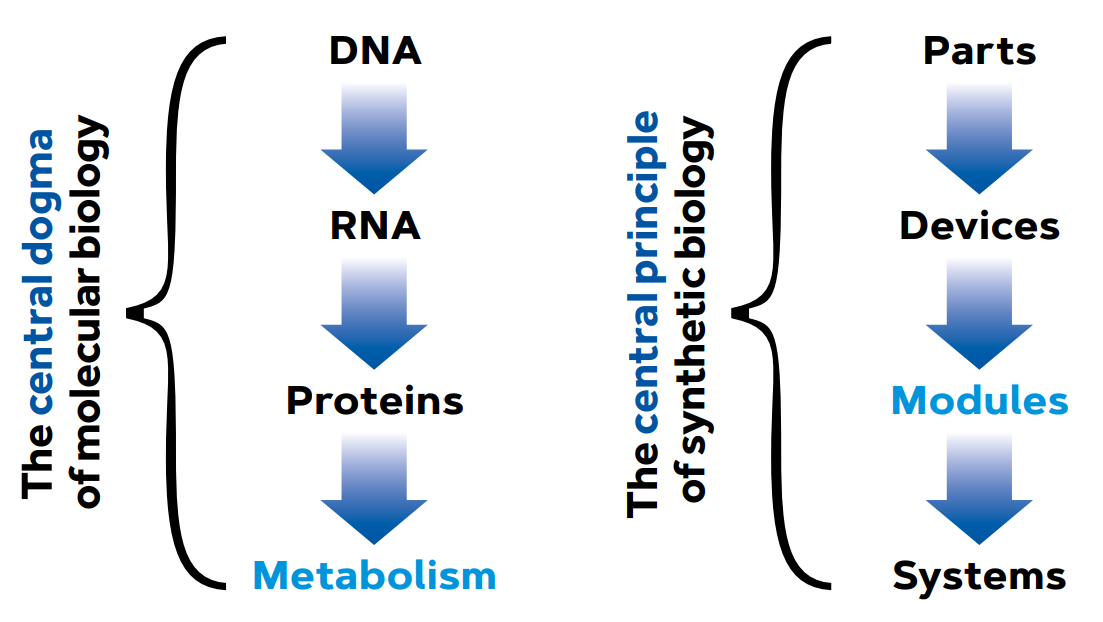
Figure 1. From the central dogma of molecular biology to the conceptual framework of synthetic biology. In the first case, the main element is the transfer of information from DNA to proteins and metabolism. Conversely, synthetic biology is concerned with the relational and compositional logic of living systems and applies typical engineering abstractions to biological objects. / Source: De Lorenzo (2018)
This «hermeneutic option» enables a vision of living systems that is compatible, yet very different, from that of molecular biology. While the latter – founded by post-war atomic scientists – promoted a way of looking at biological entities from the perspective of physics, SynBio aims to reinterpret these through the lens of engineering (Andrianantoandro et al., 2006). A side effect of this new view is that, unlike in molecular biology, the agenda of SynBio does not just involve understanding existing biological objects, but also implies their rational modification to give rise to new properties and functionalities that, in some cases, have a high economic value. In fact, one of the great frontiers in SynBio is to make the use of the term engineering – metaphorically associated with genetics during the revolution of recombinant DNA at the end of the 1970s – real in the strictest sense. Conversely, SynBio’s foundational discourse states that it is possible to move from analogy to methodology and from trial and error genetic tinkering to taking a rational approach to complex living systems, similar to the way engineers treat their technical designs (De Lorenzo, 2018; Endy, 2005). To do this, first we must deconstruct the existing biological systems into a catalogue of parts that, once standardised, can be re-assembled according to a predetermined logic to generate new functionalities.
«Synthetic biology aims to reinterpret biological entities through the lens of engineering»
Evidence for the success of this scenario is the extraordinary achievement of the so-called Registry of Standard Biological Parts, initially created and deposited at the MIT. This grew in parallel with the International Genetically Engineered Machine (iGEM) competition, which was simultaneously promoted by SynBio theorists at the same institution (Galdzicki et al., 2011). The competition, which continues to this day, was instrumental in spreading the idea of «biology-as-engineering» to numerous universities around the world and it represents an immense educational experiment that has initiated several generations of young people into the fundamental concepts of SynBio.
But let us go back to the beginning for a moment. What was the basis for the optimism expressed at the beginning by Tom Knight and his colleagues at MIT? In the years prior to the conceptualisation of SynBio, three articles appeared in the year 2000 that many consider to be foundational to the field, even though this is not explicitly expressed in any of the corresponding publications. One of these was the description by M. Elowitz’s team of the so-called repressilator: a genetic circuit between three mutually inhibitory transcriptional repressors. Under certain conditions, the repressilator results in the cyclic expression of a reporter gene – which, in this case, encoded a fluorescent protein – that acts as a visual indicator of the functioning of the system (Elowitz & Leibler, 2000; Figure 2A). The revolutionary approach of this work was that this behaviour was rationally designed following engineering principles and had a completely artificial configuration (i.e., there are no similar cases in the natural world). In spite of this, the behaviour was faithful to a mathematical model that could be simulated in a computer.
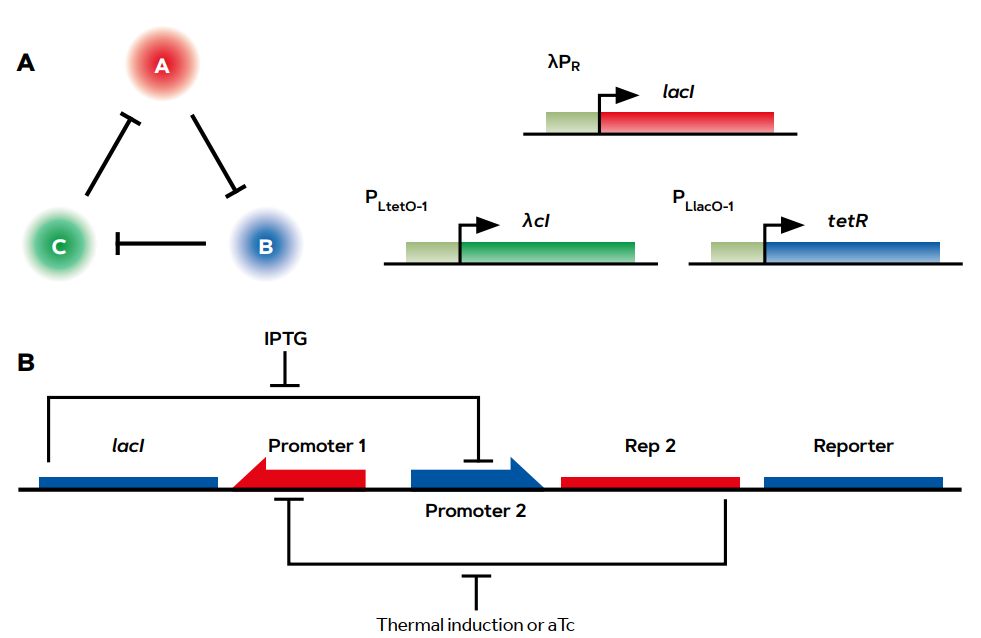
Figure 2. Two of the founding genetic constructs of synthetic biology. A) The repressilator. This genetic device is an in vivo genetic expression oscillator built with three repressors (LacI, TetR, and λCl), where each of these proteins acts as an inhibitor of the following ones by repressing their corresponding promoters (λPR, PLlacO-1, and PLtetO-1), and so each one controls the expression of the following repressor. This behaviour was rationally designed according to engineering principles and with a totally artificial configuration (there are no similar cases in the natural world). Even so, the behaviour was faithful to a mathematical model that could be simulated on a computer. B) The genetic toggle switch. This system is based on two of the previous transcriptional repressors, but is organised so they inhibit each other (LacI inhibits the tetR promoter and vice versa). This results in two stable states in which one promoter is activated while the other is deactivated. They are never active at the same time. This can be reversed by adding an inducing signal (for example, by using an IPTG reagent or anhydrotetracycline or aTc) to de-repress one of the components. This, in turn, allows the expression of the other repressor and causes a change in the state of the device. Once again, the in vivo system scrupulously obeyed the rules imposed by its human designer and responded to the predictive model. / Source: Figure A: Elowitz and Leibler (2000), Figure B: Gardner et al.
The second foundational article, published by J. Collins’s group, described the design and functioning of a genetic toggle switch (Gardner et al., 2000; Figure 2B). This was assembled with two of the biological parts used in the previous case (the transcriptional repressors) but, by wiring them in a very different way, resulted in the stable expression of two alternative genes. Again, the in vivo system scrupulously obeyed the rules imposed by its human designer and responded to the predictive model. The third publication was by L. Serrano’s laboratory and described simple genetic circuits in which negative feedback loops were rationally introduced to provide stability to the system (Becskei & Serrano, 2000). In each of the three cases, the message was the following: engineering principles can be applied to living systems, both to understand how they work and to make them work predictably in a different way. The conceptual impact of these publications was enormous, and the formulation of the discipline at MIT by Knight and his collaborators Drew Endy (Endy, 2005) and Ron Weiss (Andrianantoandro et al., 2006) can be considered the birth of modern SynBio.
The ABC of synthetic biology
The basic notion behind SynBio is that any biological system can be seen as a complex combination of independent functional elements, not unlike those found in man-made devices. On this basis, the corresponding objects can be described as the union of a limited number of components and can be rebuilt with a different configuration, either to modify existing properties or to achieve completely new ones. A fundamental aspect of SynBio is the development of material and conceptual tools for general use (biological parts, minimal genomes, artificial cells, DNA synthesis, etc.) to address problems that had been previously unsolvable. These problems include the biosynthesis of complex molecules, decomposition or recycling of toxic chemicals, biological detection of explosives, biological production of H2 and other fuels, etc. (De Lorenzo et al., 2018). But these same techniques also allow us to address completely new challenges such as DNA computing, designing developmental patterns, using bacteria to remove tumours, expanding the genetic code to non-natural amino acids, and many other surprising applications (De Lorenzo et al., 2018; O’Day et al., 2018).
«The behaviour of these biological components, regardless of their context, is a prerequisite for the engineering of new devices and properties»
One recurring premise in SynBio is the need to standardise biological components in a way that is independent from their natural circumstances. The behaviour of these components, regardless of their context, is a prerequisite for the engineering of new devices and properties. While the need and opportunity for such standardisation has been clearly identified, the success of this effort has, so far, been rather limited. Despite the long list of biological parts deposited in the MIT-backed registry, there is still a long way to go to meet the requirements that would bring them up to the standards used by the civil or electronics industry (De Lorenzo & Schmidt, 2018). This is partly because of the intrinsic property that biological functions evolved together as a whole and therefore, they behave in a highly context-dependent way. Thus, there is a need to develop better concepts and a special language to treat and classify such biological parts, based not only on their potential similarity to electronic equivalents, but also on a better understanding of their minimum biological functions, especially in relation to the regulation of gene expression (De Lorenzo & Danchin, 2008). These improvements may provide the basis for future international agreement on the format of such parts, their availability, and registration of their users.
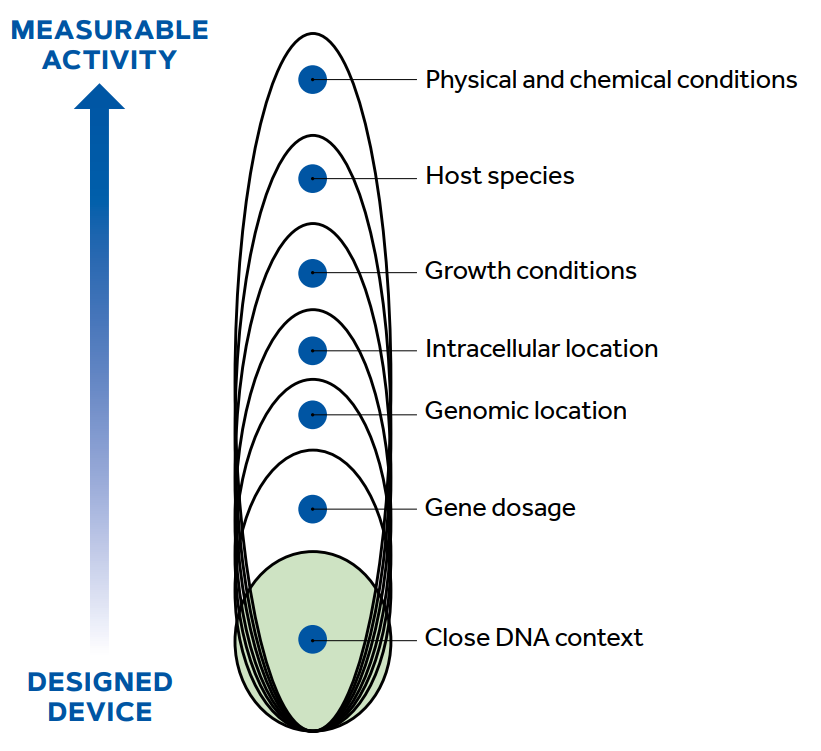
Figure 3. Contextual levels of any genetic construct. The figure summarises how the functionality of a construction must cross several layers of biological and physicochemical scenarios before the observer can measure its activity in different situations and thus, check its potential standardisation. / Source: Porcar, Danchin, and De Lorenzo (2014)
As shown in Figure 3, the functioning of each expression device in a cell is subject to at least seven contextual layers ranging from the immediate mutual influence of adjacent DNA sequences to the physical and chemical environmental conditions. It is essential to streamline the route from physical composition to final function in a measurable and quantifiable way, which requires three converging approaches. First, the detailed modelling, measurement, and parametrisation of large collections of functional parts in a variety of DNA contexts and growth conditions. Second, research into a limited number of archetypal promoters (i.e., based on consensus or very well characterised DNA sequences) in various genomic and cellular environments to identify and ultimately eliminate context-dependent determinants. Third, the advanced design of orthogonal expression devices (i.e., independent from the genetic and physiological context) which could potentially be based on biological parts recruited from mobile genetic elements (Rao, 2012). Orthogonal ribosomes and alternative genetic codes are fascinating areas of research that could also be the subject of an effort towards standardisation (Wang et al., 2007).
α and β standards to facilitate the design of living systems
In the engineering world, the terms standard and standardisation refer mainly to a) the adoption of specific geometrical shapes and size formats for the physical assembly of components in a man-made system; b) the definition of units of measurement for relevant properties and parameters, as well as the conditions and procedures necessary for calculating them (e.g., amps for current, ohms for resistance, etc.); and c) the implementation of unambiguous protocols for the manufacture of objects. These standards allow us to abstractly visualise the properties of the components of a system, accurately describe them using appropriate quantitative language (which is also standardised), and model the designed object using identical representation methods. A great advantage in this respect is the possibility of dissociating the detailed design of a product from the manufacture of its components and its final assembly. This is common in industrial engineering and electronics, but how much of this can be imported into the biological field?
«Since their inception, molecular biology and biotechnology have been affected by an almost total disregard for the challenge of standardisation»
Since their inception, molecular biology and biotechnology have been affected by an almost total disregard for the challenge of standardisation. The nomenclature for genes and molecular tools is generally chaotic. The lack of standards has been an obstacle for the comparative measurement of very basic biological functions such as promoter strength (Beal et al., 2016; Popp et al., 2017). It is therefore important to examine which of these functions can be subject to standardisation, an effort that might even lead to pre-normative action. But what can we standardise based on our current levels of knowledge?
We could distinguish between β- and α- stage standards. The former is the set of rules adopted by a certain community to improve communication, cooperation, and interoperability, but without having any pretension of universality. In this sense, the most common starting point is the establishment of a canon for the physical composition of biological devices. For example, there are now multiple formats that facilitate the assembly of DNA parts and individual modules to generate more complex systems (Casini et al., 2015). Another example is the introduction of fluorescent microspheres to calibrate and compare flow cytometry experiments (i.e., the counting and averaging of the optical emissions from individual cells in a population), as recently proposed by Beal et al. (2019). These standards are necessarily transitory because, at some point, cheaper DNA synthesis or the emergence of new platforms for measuring fluorescence will render them unnecessary.
«The adoption of formalisms derived from electrical and industrial engineering has been extraordinarily useful for the development of synthetic biology»
But there is another type of standard (which I have called α) specifically related to the metrology of the biological activities that should be developed, implemented, and promulgated. In my opinion, the most important ones are related to the measurement of the flow of gene expression from a coding DNA sequence to a protein. This flow is yet to be conceptually and materially developed. The RNA polymerase per second (PoPs) and ribosome per second (RiPS) units would be required as a universal point of reference to express the strength of the translation machinery promoter. However, these units have not yet been seriously considered, either from a fundamental point of view or as a measurement technology (Kelly et al., 2009; Sendy et al., 2016). Some argue that artificial intelligence and computer-aided design (CAD) will be able to anticipate most possible scenarios within a project on the expression of any gene of interest by compiling a large amount of experimental data on the transcription, degradation, and translation of messenger RNA (Kosuri et al., 2013). But will this be achievable in the short term? Translation is in itself a complex function that depends not only on the messenger RNA sequence, but also on its stability and a large number of physiological parameters such as cell growth phases, environmental stress, or the distribution of cellular resources. The challenges of measuring folding speed and post-translational modifications remain difficult ones. Perhaps, as is the case in other branches of engineering, future biological designs will depend entirely on automatic experience-based learning (Salis et al., 2009). But in the meantime, the rational design of living systems will be limited by the adoption of both β and α standards.

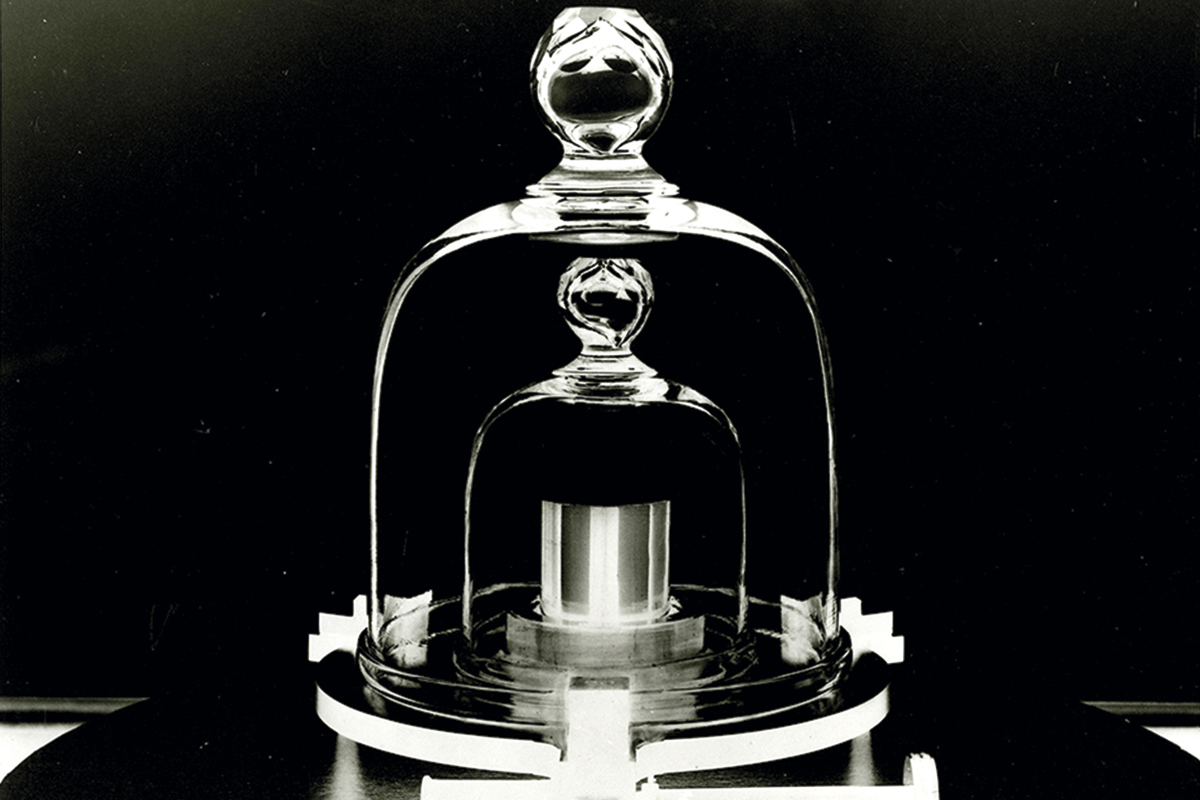
Figure 4. Standards require reference material objects. A) The metre. During the French Revolution, the National Convention had references of the new measurement installed in white marble throughout Paris. The image shows a reference installed on Vaugirard street. B) The kilo. This measurement is another product of the French Revolution. Because its definition as the weight of one litre of water was imprecise, it was redefined using a cylindrical object made of platinum and iridium as a reference. This object is kept at the International Bureau of Weights and Measures in Paris. The kilo has recently been redefined in absolute terms using quantum mechanics. The image shows a replica of the kilogram reference at the U.S. National Institute of Standards and Technology in Gaithersburg, Maryland. / Left photo: LPTP / Wikimedia Commons; right photo: National Institute of Standards and Technology Digital Collections, Gaithersburg, MD 20899
What should we do?
«To rigorously describe, measure, and “rewire” biological functions, as well as to exchange data, we must adopt standards»
Biological systems cannot automatically be equated with human artifacts. However, the adoption of formalisms derived from electrical and industrial engineering has been extraordinarily useful for the development of SynBio. The progression from parts to devices and from these to modules and systems is now a well-accepted conceptual framework in any SynBio project. But the rigorous description, measurement, and «rewiring» of biological functions, as well as the exchange data, means that we must adopt β standards in the short term, and α standards in the long term. Beyond the rules for the physical composition of DNA sequences, there is also an urgent need to develop a new type of technology which we could call in vivo biomolecular metrology. The objective of this new field would not only be the proposal of unambiguous units to describe the corresponding activities (transcription, translation, etc.), but also to generate a reference calibration entity that would allow us to coordinate measurements at different times and places. It is interesting that in the history of standards, material objects (kilos, metres, etc.) have been carefully kept as a reference for many types of measurements (Figure 4). Will it be possible to operate in the same way in the biological world?
References
Andrianantoandro, E., Basu, S., Karig, D. K., & Weiss, R. (2006). Synthetic biology: New engineering rules for an emerging discipline. Molecular Systems Biology, 2(1), 2006.0028. http://doi.org/10.1038/msb4100073
Beal, J., Farny, N. G., Haddock-Angelli, T., Selvarajah, V., Baldwin, G. S., Buckley-Taylor, R., Gershater, M., Kiga, D., Marken, J., Sanchania, V., Sison, A., & Workman, C. T. (2019). Robust estimation of bacterial cell count from optical density. BioRxiv, 803239. http://doi.org/10.1101/803239
Beal, J., Haddock-Angelli, T., Gershater, M., De Mora, K., Lizarazo, M., Hollenhorst, J., & Rettberg, R. (2016). Reproducibility of fluorescent expression from engineered biological constructs in E. coli. PLOS ONE, 11(3), e0150182. http://doi.org/10.1371/journal.pone.0150182
Becskei, A., & Serrano, L. (2000). Engineering stability in gene networks by autoregulation. Nature, 405(6786), 590. http://doi.org/10.1038/35014651
Casini, A., Storch, M., Baldwin, G. S., & Ellis, T. (2015). Bricks and blueprints: Methods and standards for DNA assembly. Nature Reviews Molecular Cell Biology, 16(9), 568–576. http://doi.org/10.1038/nrm4014
De Lorenzo, V. (2018). Evolutionary tinkering vs. rational engineering in the times of synthetic biology. Life Sciences, Society and Policy, 14(18). http://doi.org/10.1186/s40504-018-0086-x
De Lorenzo, V., & Danchin, A. (2008). Synthetic biology: Discovering new worlds and new words. EMBO Reports, 9(9), 822–827. http://doi.org/10.1038/embor.2008.159
De Lorenzo, V., & Schmidt, M. (2018). Biological standards for the Knowledge-Based BioEconomy: What is at stake. New Biotechnology, 40, 170–180. http://doi.org/10.1016/j.nbt.2017.05.001
De Lorenzo, V., Prather, K. L. J., Chen, G.-Q., O’Day, E., Kameke, C., Oyarzún, D. A., Hosta-Rigau, L., Alsafar, H., Cao, C., Ji, W., Okano, H., Roberts, R. J., Ronaghi, M., Yeung, K., Zhang, F., & Lee, S. Y. (2018). The power of synthetic biology for bioproduction, remediation and pollution control: The UN’s Sustainable Development Goals will inevitably require the application of molecular biology and biotechnology on a global scale. EMBO Reports, 19(4), e4658. http://doi.org/10.15252/embr.201745658
Elowitz, M. B., & Leibler, S. (2000). A synthetic oscillatory network of transcriptional regulators. Nature, 403(6767), 335–338. http://doi.org/10.1038/35002125
Endy, D. (2005). Foundations for engineering biology. Nature, 438(7067), 449–453. http://doi.org/10.1038/nature04342
Galdzicki, M., Rodriguez, C., Chandran, D., Sauro, H. M., & Gennari, J. H. (2011). Standard biological parts knowledgebase. PLoS ONE, 6(2), e17005. http://doi.org/10.1371/journal.pone.0017005
Gardner, T. S., Cantor, C. R., & Collins, J. J. (2000). Construction of a genetic toggle switch in Escherichia coli. Nature, 403(6767), 339–342. http://doi.org/10.1038/35002131
Kelly, J. R., Rubin, A. J., Davis, J. H., Ajo-Franklin, C. M., Cumbers, J., Czar, M. J., de Mora, K., Glieberman, A. L., Monie, D. D., & Endy, D. (2009). Measuring the activity of BioBrick promoters using an in vivo reference standard. Journal of Biological Engineering, 3(1), 4. http://doi.org/10.1186/1754-1611-3-4
Kosuri, S., Goodman, D. B., Cambray, G., Mutalik, V. K., Gao, Y., Arkin, A. P., Endy, D., & Church, G. M. (2013). Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proceedings of the National Academy of Sciences, 110(34), 14024–14029. http://doi.org/10.1073/pnas.1301301110
O’Day, E., Hosta-Rigau, L., Oyarzún, D. A., Okano, H., de Lorenzo, V., von Kameke, C., Alsafar, H., Cao, C., Chen, G.-Q., Ji, W., Roberts, R. J., Ronaghi, M., Yeung, K., Zhang, F., & Lee, S. Y. (2018). Are we there yet? How and when specific biotechnologies will improve human health. Biotechnology Journal, 14(1), e1800195. http://doi.org/10.1002/biot.201800195
Popp, P. F., Dotzler, M., Radeck, J., Bartels, J., & Mascher, T. (2017). The Bacillus BioBrick Box 2.0: Expanding the genetic toolbox for the standardized work with Bacillus subtilis. Scientific Reports, 7(1), 15058. http://doi.org/10.1038/s41598-017-15107-z
Porcar, M., Danchin, A., & De Lorenzo, V. (2014). Confidence, tolerance, and allowance in biological engineering: The nuts and bolts of living things. Bioessays, 37(1), 95. http://doi.org/10.1002/bies.201400091
Rao, C. V. (2012). Expanding the synthetic biology toolbox: Engineering orthogonal regulators of gene expression. Current Opinion in Biotechnology, 23(5), 689–694. http://doi.org/10.1016/j.copbio.2011.12.015
Salis, H. M., Mirsky, E. A., & Voigt, C. A. (2009). Automated design of synthetic ribosome binding sites to control protein expression. Nature Biotechnology, 27(10), 946–950. http://doi.org/10.1038/nbt.1568
Sendy, B., Lee, D. J., Busby, S. J., & Bryant, J. A. (2016). RNA polymerase supply and flux through the lac operon in Escherichia coli. Philosophical Transactions of the Royal Society B: Biological Sciences, 371(1707), 20160080. http://doi.org/10.1098/rstb.2016.0080
Wang, K., Neumann, H., Peak-Chew, S. Y., & Chin, J. W. (2007). Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nature Biotechnology, 25(7), 770–777. http://doi.org/10.1038/nbt1314