The genome and the complexity of living beings. The genome of an organism is the whole DNA content of its cells, including genes and intergenic regions. In prokaryotes (Archaea and Bacteria) there is, in general, a linear relationship between genome size and the number of genes. The smallest genomes are found in symbionts and parasites, as they undergo a gene degradation process during adaptation to their new lifestyle. However, in eukaryotes there is no correlation between genome size and the complexity of the organism. This is known as the C-value paradox. The largest genome is found in an amoeba, a one-cell organism, with 686,000 Mb, 200 fold larger than the human genome and 20,000 fold larger than the one found in yeast. Now we know that most excess DNA is repetitive DNA, apparently lacking a function (selfish DNA) and whose possible role in genome evolution is still unknown.
As we know, DNA is the material of genes; therefore, it is natural to think that these more complex organisms will require more genes and will have more DNA. Accordingly, one might expect that: «more complex organisms have larger genomes and contain a larger number of genes». That is, throughout evolution an increase in genome sizes and the number of genes is expected.
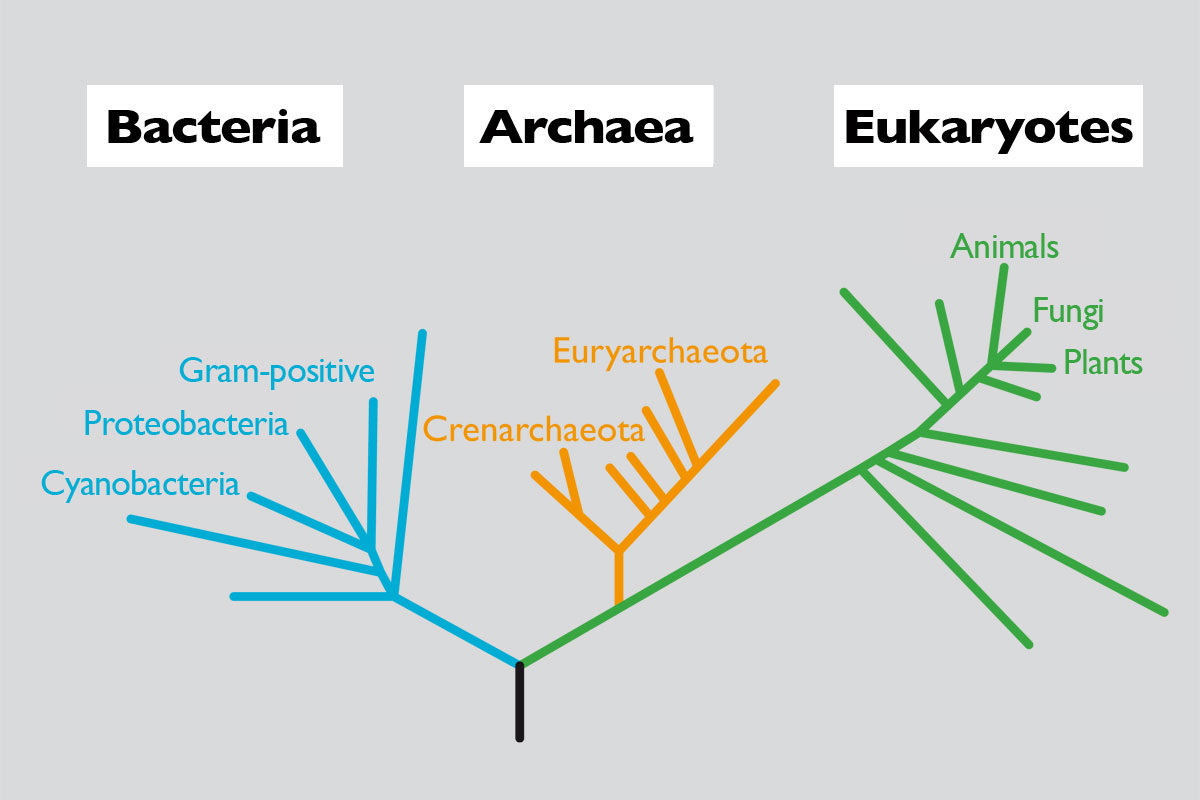
To begin the study of genome sizes, and try to see if our expectations match the observations, we will adopt the classification of life on earth as proposed by Woese et al (1990) following phylogenetic studies of the 16S rDNA gene, which codes for the small subunit of ribosomal RNA. This is a gene highly conserved in the evolutionary scale and that seems to reproduce well the relationship between living beings. According to the phylogeny obtained, the authors propose that cellular life on earth can be grouped into three domains: bacteria and archaea (both prokaryotes) and eukaryotes. A simplified relationship can be seen in Figure 1.
GENOME RANGES AND SIZES
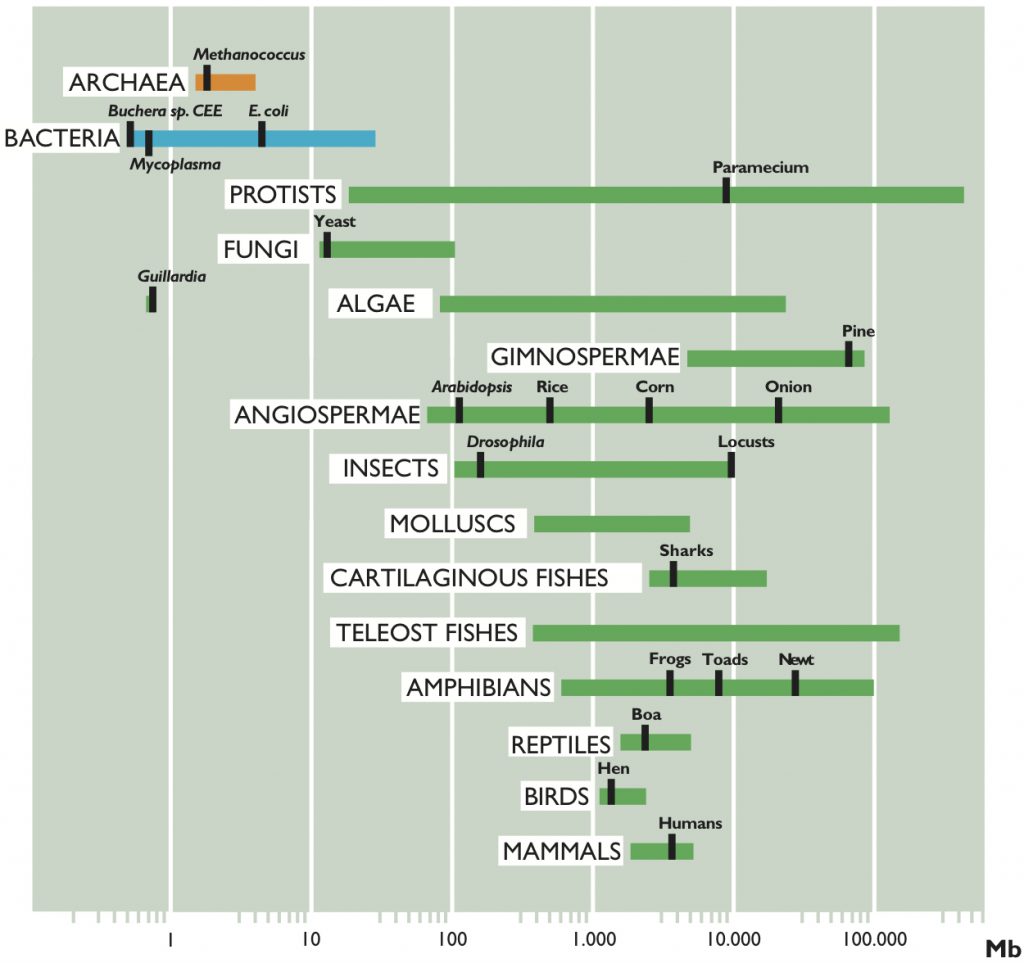
Figure 2 represents the range of sizes of genome found in the three domains of life: bacteria, archaea and eukaryotes. It seems clear that prokaryotes are, in general, smaller than eukaryotes, with the exception of some large-sized bacteria and some very small-sized eukaryotes. Let us see the data in more detail.
PROKARYOTES: BACTERIA AND ARCHAEA
According to the data published so far, the size varies from 0.58 megabases (1 megabase (Mb) is one million base pairs (bp)) in the intracellular pathogen Mycoplasma genitalium, to more than 10 Mb in several species of cyanobacteria, with the exception of Bacillus megaterium, which has a genome of 30 Mb. The second smallest genome ever published is that of Buchnera sp. APS, endosymbiont of the cereal aphid Acyrthosiphon pisum, with a size of 641 kb. Recently, our research group has characterized six genomes smaller than even those of Mycoplasma, the smallest of all being that of Buchnera sp. CCE, endosymbiont of the aphid Cinara cedri, with a size of 0.45 Mb. In general, most genomes are less than 5 Mb in size, as shown in Table 1.
Is there a relationship between genome size and number of genes? The size of the prokaryotic gene is uniform, about 900 to 1000 bp. Therefore, one can estimate the gene density at each sequenced genome. As seen in Table 1, gene density is more or less constant, both in bacteria and archaea.
We can conclude that, at least in prokaryotes, genomes have a larger number of genes and are also more complex. That is, the number of genes reflects the lifestyle. Thus, smaller bacteria are specialists, such as obligate parasites and endosymbionts, and larger bacteria are generalists, and may even have a certain degree of development, such as sporulation in Bacillus.
EUKARYOTES: C-VALUE PARADOX
Genome size in eukaryotes is defined as the C-value or amount of DNA per haploid genome, such as that which exists in the nucleus of a spermatozoon. It is called C, for constant or characteristic, to indicate the fact that size is practically constant within a species.
Referring back to Figure 2, we see that, in general, eukaryotes have larger genomes than prokaryotes, except for some endosymbiont or parasitic green algae, which have very small genomes. Specifically, the smallest eukaryotic genome ever sequenced is that of Guillardia theta, a symbiont red algae, of only 0.55 Mb. We can also see in the figure that there is a wide range of sizes, much greater than that of prokaryotes, more than 80,000-fold larger, from organisms such as yeast (1.2 Mb) to the amoeba (686,000 Mb). But is there, as in bacteria, a relationship between genome size and complexity of the organism?
In Figure 2 we have represented the range of C-value in several representative groups of eukaryotic organisms. As we can observe, unicellular protists such as amoebae show the greatest variation in C-values (23.5 Mb to 686.000 Mb, with a ratio of 29,191 between the largest and the smallest), while mammals, birds and reptiles show less variation in the size of their genome (a ratio of only 4, 1 and 4, respectively). Furthermore, the large variation in genome sizes between eukaryotic species does not seem to have a relationship with either the complexity of the organism or the number of genes they contain. For example, amoebae, which have the largest genomes, have 200 times more DNA than humans (3,400 Mb) and it is clear that an amoeba cannot be more complex than a human. Moreover, it would be expected that mammals, more complex organisms, present larger genomes. However, many other organisms, such as fish, amphibians or plants, have much larger genomes. Even when we compare the sizes between organisms that appear similar in terms of complexity, there are also wide differences in their C-values. To give some examples, flies and locusts, onions and lilies, etc. have considerable variations in the sizes of their genomes. Amphibians as a group have variations of up to 91 times and it is hard to believe that this may reflect variations of nearly 100 times the number of genes necessary to give rise to the corresponding amphibians, or that onions need 200 times more DNA than rice. Figure 3 shows some living beings with size proportional to the size of their genome and needs no further explanation.
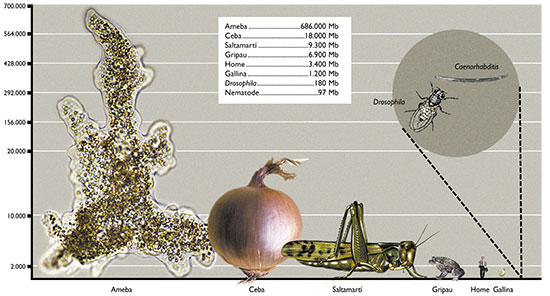
Figure 3. Genome size in some living beings. The height of the drawings is proportional to the size of their genome. The specimes are amoebae, onions, grasshoppers, toads, humans, hens, Drosophila and Caenorhabditis, a nematode worm.
The mismatch between the C-values and the presumed amount of genetic information contained within the genomes was called C-value paradox. Since we cannot assume that a species possesses less DNA than the quantity required to specify its vital functions, we have to explain why many species contain this amount of excess DNA.
GENE DNA OR NON-GENE DNA
The first question that has to be clarified is whether there is a correlation between genome size and the number of genes. That is, are the differences in genome sizes due to gene or non-gene DNA?
We have known since the late 60s that the eukaryotic genome is composed of a large amount of repetitive DNA. Moreover, since the late 70s we have known that genes are interrupted by non-coding sequences, introns, which must be removed before the ribosome synthesizes protein. We are talking in both cases about a seemingly superfluous DNA, which contributes to the wide variation in C-values and therefore explains the apparent paradox.
The size and number of introns vary widely along the evolutionary scale, mammals being the ones with the highest number and larger size. Repetitive DNA also varies between organisms. Traditionally this DNA is classified as: highly repetitive, with sequences such as microsatellites and minisatellites; and moderately repetitive, where transposable elements, the sequences that constitute the clearest example of selfish DNA, are found.
Number of genes and complexity of the organism
As sequences of whole genomes are completed, we will know with more or less accuracy the number of genes derived from these sequences, since what we had so far were indirect estimates. However, some data is proving to be surprising because, in some cases, there appears to be a clear correlation between the number of genes and the complexity of the organism. The nematode worm C. elegans has 18,000 genes (Table 1), about 5,000 more than Drosophila, a more complex organism. Man has only twice as many genes as C. elegans (estimates indicated about 100,000). We are also beginning to understand these data. There are mechanisms in higher eukaryotes that are able to «expand the proteome». That is, from the same DNA sequence, they can obtain more than one protein. Large introns found in mammals can, in many cases, «hide» information that cannot be inferred only with the DNA sequence. It will be some time before we can determine the number of proteins that an organism is able to synthesize. But this would be the subject of another paper. In any case, we can say that «more complex organisms have more gene functions.»
| Organism | ||||
| Common name or class | Scientific name | Genome size (Mb) |
Number of genes | Gene density (genes/Mb) |
| Eukaryotes | Saccharomyces cerevisiae | 12 | 6,241 | 480 |
| Baker’s yeast | ||||
| Nematode | Caenorhabditis elegans | 97 | 18,424 | 190 |
| Cruciferous | Arabidopsis thaliana | 125 | 25,498 | 204 |
| Fruit fly | Drosophila melanogaster | 180 | 13,601 | 75 |
| Pufferfish | Fugu rubripes | 400 | 35,000 | 100 |
| Rice | Oryza sativa | 450 | ||
| Sea urchin | Strongylocentrotus purpuratus | 900 | 27,350 | 30 |
| Maize | Zea mays | 2,400 | ||
| Human | Homo sapiens | 3,400 | 35,000 | 10 |
| Onion | Allium cep | 18,000 | ||
| Amoeba | Amoeba dubia | 686,000 | ||
| Archaea | Aeropyrum pernix | 1.55 | 1,522 | 981 |
| Crenarchaeota | ||||
| Euryarchaeota | Methanococcus jannaschii | 1.66 | 1,715 | 1033 |
| Euryarchaeota | Archaeoglobus | 2.18 | 2,420 | 1110 |
| Bacteria | Buchnera sp. CCE | 0.45 | ||
| Proteobacteria | ||||
| Gram positive | Mycoplama genitalium | 0.58 | 479 | 831 |
| Proteobacteria | Buchnera sp. APS | 0.64 | 564 | 881 |
| Gram negative | Haemophilus influenzae | 1.8 | 1,727 | 959 |
| Cyanobacteria | Synechocystis sp. | 3.6 | 3,168 | 880 |
| Gram positive | Bacillus subtilis | 4.2 | 4,100 | 976 |
| Proteobacteria | Escherichia coli | 4.6 | 4,288 | 932 |
Table 1. Genome size, gene number and gene density.
MOLECULAR MECHANISMS THAT ALTER GENOME SIZE
There are many mutational mechanisms that can produce changes in genome size. Some of them occur on a large scale (whole genome duplication), while others occur on a very small scale (loss or gain of a few nucleotides). However, we must note that these mutations affect, in principle, a single cell, and if it is a gamete, the mutation may be transmitted to an offspring. This individual will have to live with others in a population and only in future generations will we know if the mutation it carries will extend to all individuals in the population (fixation) or, on the contrary, will disappear. The probability of one or the other occurring depends on evolutionary mechanisms such as natural selection (whether it provides an advantage or disadvantage to the individual) or genetic drift (random).
Chromosomal mechanisms often produce drastic changes with a single mutation. We can highlight the whole genome duplication, duplication affecting a single chromosome, or part of it. Equivalently, mutations that result in the loss of some chromosome fragment are also known. Such changes, though also frequent in plants and animals, appear to have significantly contributed to shape the evolution of the genome of the former.
«We can say that “more complex organisms have more gene functions”»
The mobile genetic elements or transposable elements, are other causes of large variations in genome size. These elements, of a few thousand nucleotides, are duplicated and the duplicate copies are inserted in other parts of the genome, causing rapid increases in its size, unless the mechanisms that control their proliferation intervene (as authentic selfish elements, they develop mechanisms to regulate their movement in the genome, so as not to produce irreparable damage, since that would imply their own disappearance). It has been estimated that the maize genome has been duplicated because of the transposable elements in the last three million years of its evolution.
It is considered that the spontaneous insertions or deletions (called indels) of a few nucleotides are one of the most important causes of the development of the size of the genome on the long term. In several species of insects, for example, a strong correlation has been shown between the overall rate of DNA loss of intergenic and non-coding regions and genome size. The fixation of these mutations is very unlikely if the indel affects a gene, but it is more likely if it affects pseudogenes (nonfunctional genes, recently inactivated) or other nonfunctional DNA sequences. The disintegration of the genes, or the disappearance of a gene in the genome, usually occur in a first step with its inactivation by a point mutation (formation of a pseudogene). Subsequently, the DNA that forms the pseudogenes is progressively removed until it disappears completely from the genome.
The variation in the length of the DNA of the minisatellites and microsatellites is another mechanism which can alter the size of the genome. These sequences are formed by a unit of few nucleotides, repeated contiguously from less than ten to several thousand times. The number of repeated units varies greatly, even in individuals of the same species. This is due to two mechanisms that cause both an increase and a decrease in the number of repetitions. These mechanisms are the recombination, called unequal, and the errors during DNA replication, due to the phenomenon known as slippage of the DNA polymerase. Indeed, these are the sequences that are being used for the identification of human remains, for example, due to their high variability.
One of the most important processes in the increase of genome size in unicellular organisms, especially prokaryotes, is horizontal gene transfer. This process involves the introduction into the genome of a DNA fragment from another species, containing one or more genes. The DNA is introduced into the cell by different mechanisms and must then recombine with the genome. If the genes introduced confer some advantage to the organism they remain like that, otherwise they can mutate and become inactive. An example of the importance of these processes can be seen in the intestinal bacteria Escherichia coli, some strains of which have become pathogenic for the transfer of virulence genes. The significance of this phenomenon in recent periods of evolution of higher organisms is much more limited, but there are many reported cases of intracellular DNA gene transfer from the genome of the mitochondria and chloroplasts to the nuclear genome.
All these mutations arise periodically in many species, including humans. Many of them are disadvantageous and selective pressure progressively removes them from the population, others are individually neutral but may be collectively beneficial or harmful, by increasing or decreasing the size of the genome. In bacteria, having a small genome can be positive to optimize the time and cost of DNA replication. In eukaryotic organisms, several advantages have been proposed for having a large genome size, though this is not always associated with a higher number of genes.