Beyond the double helix
Structure of DNA G-quadruplexes
DNA is the fundamental biomolecule required for correct cell functioning and, until very recently, it was associated with the double helix structure discovered over 70 years ago by Crick, Watson, and Franklin. However, other DNA structures and conformations such as G-quadruplexes have also been described. These G-quadruplexes are formed in genomic regions that are rich in guanine. They have a tetramer structure and control biological processes such as gene expression, protection against ageing, or the transmission of neural information. In this document, we describe their chemical and structural characteristics and discuss their main cellular functions. Lastly, we present G-quadruplexes as potential molecular targets for future cancer therapies.
Keywords: G-quadruplexes, DNA, telomeres, oncogene, drug.
Today it is well-known that nucleic acids are fundamental biomolecules for correct cellular functioning and in the regulation of biological processes. These biomolecules can fold into several structures and conformations. However, the most well-known structure is the one obtained by Rosalind Franklin, James Watson, and Francis Crick in the 1950s: a double helix of deoxyribonucleic acid (DNA). Discovering the structure of the molecule of life revolutionised 20th-century molecular biology because it helped scientists to understand how genetic information was stored, protected, and transmitted. DNA is a helical polymer structure composed of four nucleotides that differ from each other in the nitrogenous base they contain. These nucleotides are adenine (A), guanine (G), thymine (T), and cytosine (C). Thus, DNA can be understood as a letter code guiding the construction of ribonucleic acid or RNA. In turn, RNA is mainly used to generate the proteins that regulate all the biological functions and characteristics of living beings, such as eye colour, hair type, or hand size.
Apart from the double helix structure, DNA has been shown to create other alternative conformations that control different biological functions, such as gene expression or protection against ageing. Among these structures, G-quadruplex DNA (G4) has received the most attention over the last 10 years because of its role as an epigenetic regulator and as a therapeutic target for cancer and other diseases. In this article, we want to introduce readers to this new DNA structure and its structures and molecular and biological functions. In addition, we analyse its potential applications as a therapeutic target for several diseases and explain the future research perspectives in this field.
The structural diversity of DNA
The race to discover the structure of DNA culminated with Rosalind Franklin’s fibre diffraction experiments, which resulted in the famous X-ray Photo 51. Based on that photograph, James Watson and Francis Crick published a description of the double helix structure in Nature (Watson & Crick, 1953) and years later, in 1962, they were awarded the Nobel Prize in Physiology and Medicine, together with Maurice Wilkins (Oregon State University, 2015). This discovery became the most significant and extraordinary advance in molecular biology and one of the most important scientific breakthroughs of the 20th century.
The double helix structure consists of two antiparallel chains coiled around each other forming a spiral and thereby creating a helical structure. As mentioned above, DNA strands are formed from four different units, called nucleotides. Nucleotides include a phosphate group, a sugar group, and a nitrogenous base. The nitrogenous bases are adenine, guanine, thymine, and cytosine. The phosphate groups face the outside of the structure, while bases are located on the inside and result in pairs of facing bases (guanine–cytosine and adenine–thymine) connected by hydrogen bonds. Each base pair is placed on top of the previous one, with a fixed rotation. Thus, a spiral is centred on the main axis of the structure. Many artists, sculptors, and architects have used this shape in their works. In addition, each nucleic acid strand has two ends called the 5’-end and 3’-end, which we use to assign a polarity or directionality when describing DNA processes.
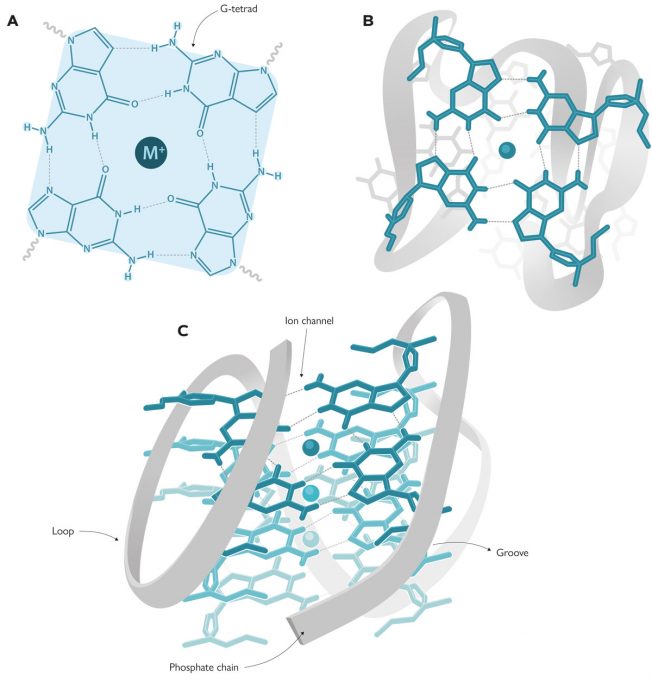
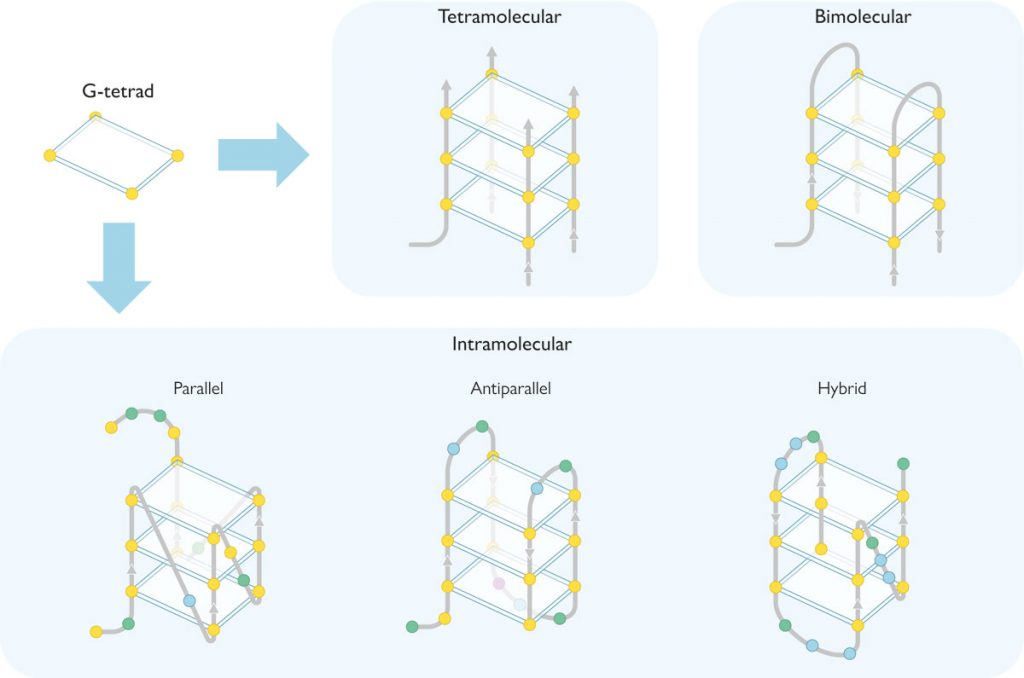
However, the double helix is not the only DNA structure we know of. There are other non-canonical forms including triplex DNA (triple-stranded DNA), i-motif DNA (similar to G-quadruplexes), or replication forks, which can allow DNA molecules to take on a multitude of conformations. In fact, about 10 years after the DNA structure was deciphered, Martin Gellert, David R. Davies, and their colleagues published the first guanine gel structures forming G-tetrads (Gellert et al., 1962), in other words, the basic units for G-quadruplexes. These G-tetrads, also known as G-quartets, contain four guanines connected with hydrogen bonds in a planar array (Figure 1A–B). But it was not until the 1980s that Elizabeth H. Blackburn and her team described the first G-quadruplex structure formed by DNA sequences in telomeres (Henderson et al., 1987). This discovery aroused great interest in the scientific community, not only because of the structure itself, but also because of its biological implications. This led to a rapid increase in the number of studies focused on discovering which parts of the human genome could form these structures. Using bioinformatics techniques and by analysing genome sequences, researchers located 700,000 regions in the human genome that could form G-quadruplexes (Chambers et al., 2015). But the most extraordinary discovery was that their locations were not random, as would be expected from a non-functional region. These regions were detected mainly in telomeres and oncogene promoters, indicating that G-quadruplexes play a paramount role in living beings. In the words of Aaron Klug, who received a Nobel Prize in Chemistry for the development of crystallographic methods to decipher protein-nucleic acid complexes, «if G-quadruplexes form so readily in vitro, Nature will have found a way of using them in vivo».
G-quadruplex structure
As noted above, G-quadruplexes are formed by rings of four guanines connected with hydrogen bonds in planar array, known as G-tetrads. Each G-quadruplex contains two or more G-tetrads stacked like floors in a building, surrounding a sort of internal conduct (an ion channel) containing cations, usually sodium or potassium (Figure 1). Depending on the size of the cation, it can be located on the «same floor of the building» (i.e., on the same plane as each G-tetrad) and will form four bonds with the guanines, as is the case with sodium. However, potassium is larger and is located «between two floors» (i.e., between two G-tetrads) where it forms eight bonds (Neidle & Balasubramanian, 2006).
G-quadruplexes can be found in a multitude of conformations governed by different parameters. Thus, they can consist of one (intramolecular quadruplex), two (bimolecular), or four (tetramolecular) guanine-rich DNA strands (Figure 2). Depending on the relative orientation of these strands, they can also take several conformations: parallel, antiparallel, and hybrid (Figure 2). In the parallel conformation, all the strands have the same orientation, whereas in antiparallel conformations, the orientation is opposed for two of them. In hybrid conformations, three strands proceed in one orientation, while the rest go in the opposite orientation. Finally, in G-quadruplexes these DNA strands form loops and grooves (Figure 1C), which depend on the composition of the nucleotides in the strands and other external factors such as ionic strength or temperature.
The G-quadruplex in telomeres
Telomeres are located at the ends of chromosomes and consist of regions of non-coding, highly repetitive DNA sequences. Their function is essential to maintain chromosome integrity in processes such a gene recombination, chromosome fusion, and nuclease degradation. In short, telomeres preserve genetic information during cellular division and thus preserve the integrity and functionality of genes over time. More specifically, telomeric DNA consists of a repetition of guanine-rich nucleotides – usually TTAGGG for eukaryotic cells – between 9,000 and 15,000 nucleotides long. Although telomeric DNA has a double helix conformation, the 3’-end always contains an extra single-stranded DNA fragment which is 50–300-bases long that is not required by the complementary chain (Figure 3).
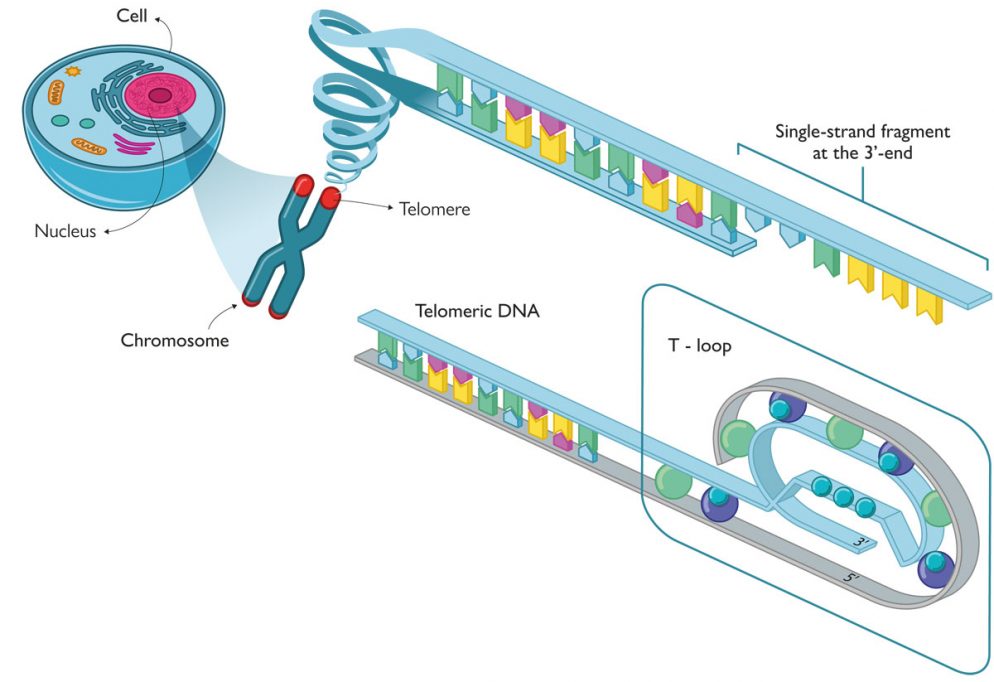
The single-stranded DNA in telomeres forms a loop structure (known as a T-loop) in which the DNA coils around itself and to which a number of proteins is also attached. This protein structure is called the shelterin supercomplex or telosome and protects telomeres from the usual DNA degradation (end unpairing) and is found in single strands that are unpaired with their complementary chain (Figure 3). Despite the important role of telomeres in chromosome protection, they are shortened with every cell division which is known as the «end replication problem», which eventually leads to cellular death. According to the typical DNA replication mechanism, the polymerase enzyme requires a small fragment of DNA, called a primer, to trigger the addition of nucleotides to the newly formed DNA strand. Because the addition always goes from the 5’-end to the 3’-end, one of the two strands is continuously synthesised, while the other has to be built discontinuously with short DNA fragments known as Okazaki fragments. Each of these fragments has its own primer, which is used as a template to start adding nucleotides from the 5’-end to the 3’-end. At the end of the process, RNA fragments are replaced with DNA, with the exception of the last primer, the one for the 5’-end. This is precisely «the problem» and the origin of the single-stranded 3’-end fragment on telomeres. Consequently, each cell cycle involves a net loss of between 100 and 200 nucleotide bases. After a certain number of cycles, what is known as the Hayflick limit is reached, and telomeres are so short that the T-loop cannot be formed and further cycles are no longer possible. At this point, the cell becomes very genomically instable and enters senescence; cell division then stops and ultimately results in cell death.
Thus, telomere length is associated with ageing and the biological age of cells, which may be different from the chronological age of the individual. Therefore, cells with long telomeres are considered young and can lead to a high number of divisions, while cells with short telomeres duplicate less frequently and are closer to dying. There is a direct relationship between telomeres and some lifestyle aspects such as stress levels or unbalanced diets. They are also strongly associated with age-related diseases such as cancer and cardiovascular or neurodegenerative diseases like Alzheimer’s. This phenomenon also arouses great interest among the general public. In fact, some books provide advice on how to maintain telomeric length in order to improve one’s quality of life. There are even companies like LifeLength® or TeloYears® dedicated to measuring telomere length to estimate one’s real biological age.
Nonetheless, biological systems have developed a general mechanism to address the problem of telomeric shortening: the telomerase enzyme. This enzyme is responsible for maintaining telomere length and can synthesise DNA using an RNA template. We owe its discovery to Carol Greider, Elisabeth Blackburn, and Jack Szostak, who were awarded the Nobel Prize in Medicine for their discovery in 2009. Telomerase is an enzyme complex formed by several components: different protein groups, a reverse transcription protein, and an RNA template, complementary to the 3’-end of the telomere. Several factors are involved in regulating this process, and telomerase is not active in all cells. More specifically, most cells in a living being are somatic and experience progressive shortening of their telomeres, while certain cells such as embryonic cells, stem cells, and epithelial cells have an active telomerase.
However, telomerase is overexpressed in approximately 90 % of the cells related to the uncontrolled proliferation of tumours and the immortalisation of those cells. Immortalised cells divide endlessly because they have evaded natural cell death mechanisms. This is the case with cancer cells whose proliferation, unlike regular cells, is almost unlimited. Precisely for this reason, telomerase is considered a compelling therapeutic target for cancer treatment. In fact, there is currently an extensive line of research focused on designing molecules to act as telomerase inhibitors – by stabilising G-quadruplex structures in telomeres – to slow down tumorigenic processes.
G-quadruplexes in oncogene promoters
DNA sequencing and bioinformatics analyses have allowed us to locate G-quadruplex-forming sequences in the promoters of a multitude of genes that produce cancer (genes otherwise known as oncogenes). Unlike telomeres, promoters are always double-stranded DNA fragments, which a priori, suggests that it would be more difficult to them to form the G-quadruplex structure. But gene transcription processes require the previous unpairing of the double helix, which facilitates the formation of G-quadruplexes. Once the DNA is unpaired, the G-quadruplex structure can be formed from a guanine-rich strand. This can occur through the action of certain proteins and small molecules that regulate oncogene expression (Danzhou & Keika, 2010). Some molecules have been shown to stabilise G-quadruplexes in oncogene promoters. Thus, they represent an obstacle to transcription, including the transcription of oncoproteins, which can ultimately lead to novel treatments for cancer. Therefore, another strategy for cancer treatment is the formation of G-quadruplexes in oncogene promoters by using small molecules obtained in the laboratory.
Many G-quadruplex-forming sequences have been found in oncogene promoters, even though most research attention has focussed on only three of them because of their importance in the onset and development of cancer. These three are the c-Myc transcription factor oncogenes, Bcl-2 mitochondrial protein regulators, and the c-Kit receptor tyrosine kinase.
The first oncogene identified with a G-quadruplex in its promoter sequence was c-Myc. The c-Myc protein contributes to the regulation of approximately 15 % of the genes in the human genome; therefore, it is closely related to apoptosis or programmed cell death, metabolism, proliferation, and general control of the cell life cycle. When c-Myc is overexpressed, these regulation processes become altered and other genes promoting cancers such as breast, colon, or brain cancer are activated. The DNA region corresponding to the c-Myc promoter contains a sequence with 27 guanine-rich bases, which facilitate the formation of a parallel G-quadruplex structure. We know that this region controls the transcriptional activity of c-Myc and, as we mentioned above, the formation of G-quadruplexes represents an obstacle for oncogene transcription.
On the other hand, the Bcl-2 gene has a guanine-rich region in its promoter that also regulates its expression and can form a hybrid G-quadruplex structure with an exceptionally long loop. The protein product of the Bcl-2 oncogene is a mitochondrial protein related to the control of cellular death because it functions as an apoptosis inhibitor for cancer cells. This protein is overexpressed in many cancers including breast, prostate, lung, colon, cervical, or lymphatic cancers. Thus, stabilising the G-quadruplex in the Bcl-2 promoter could induce apoptosis and, consequently, be useful to cure cancer.
«Several G-quadruplex molecules – either from natural extracts or synthesised in the laboratory – seem to be very promising for cancer therapies»
As for the c-kit gene, its promoter presents two regions rich in guanine, known as c-kit1 and c-kit2. These sequences are 22 and 20 bases long, respectively, and are located close to where the oncogene transcription starts and, therefore, are related to the onset of cancer. They can both form different types of G-quadruplex structures. The c-kit1 sequence forms a parallel quadruplex with a large loop stabilised by the interactions between base pairs of the same sequence. Conversely, the G-quadruplex in c-kit2 shows a more conventional parallel topology. In this case, c-kit encodes a transmembrane receptor that stimulates cellular proliferation and differentiation. This gene has proved to be closely related to many tumour types such as those affecting the lymph nodes or lungs and, especially, gastrointestinal cancer. Since the c-kit promoter regions rich in guanine are essential for its transcription, stabilisation and formation of its G-quadruplexes must play an essential role in inactivating the expression of tumour cells.
G-quadruplexes are also formed in RNA
All of the examples mentioned so far correspond to G-quadruplex DNA, but RNA can also result in tetramer G-quadruplex structures. Because it is single-stranded, RNA has a much greater capacity than DNA to form secondary non-canonical structures in vivo. As in the case of quadruplex DNA, the existence of messenger RNA sequences forming quadruplex structures in cells have also been proven. The formation of quadruplex RNA (or lack thereof) alters the cellular transcriptome, thus impacting their biological functions. However, RNA G-quadruplex research has just begun and so which functions it relates to still remains unknown.
The future of G-quadruplexes
DNA is now one of the main molecular targets for cancer therapies. This is the case, for example, of cisplatin, which accounts for more than 90 % of current oncological treatments. Platinum-based drugs such as this interact with the double helix of DNA but do not discriminate between tumour and non-tumour cells, thereby resulting in many adverse side effects and the potential for patients to acquire resistance to them. Therefore, much of today’s research focuses on the design of new drugs with alternative therapeutic targets such as G-quadruplex DNA. As mentioned above, these intriguing structures are formed in very specific locations in the genome, such as telomeric or oncogene promoter DNA.
The pharmaceutical industry and academic laboratories have now developed several G-quadruplex molecules – either from natural extracts or synthesised in the laboratory – which seem to be very promising for cancer therapy (Neidle, 2017; Pont et al., 2020). In clinical trials, some molecules such as quarfloxin have proven to efficiently target liver cancer, one of the most lethal and aggressive tumour types. Quarfloxin forms and stabilises a ribosomal G-quadruplex that inhibits the prolongation of polymerase and has powerful antitumor activity.
On the other hand, some studies highlight the important role of G-quadruplex structures in different biological processes, although their exact mechanisms and repercussions have not yet been fully defined. For instance, we know that G-quadruplexes also have implications in different neurodegenerative processes. Consequently, a large number of research groups are focusing on the design of treatments for these diseases, using quadruplexes as a regulator. In conclusion, an exciting age is coming for research on how these G-quadruplex structures truly work. They may represent key targets for the treatment of diseases such as cancer or neurodegenerative pathologies.
References
Chambers, V. S., Marsico, G., Boutell, J. M., Di Antonio, M., Smith, G. P., & Balasubramanian, S. (2015). High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nature Biotechnology, 33(8), 877–881. https://doi.org/10.1038/nbt.3295
Danzhou, Y., & Keika, O. (2010). Structural insights into G-quadruplexes: Towards new anticancer drugs. Future Medicinal Chemistry, 2(4), 619–646. https://doi.org/10.4155/fmc.09.172
Gellert, M., Lipsett, M. N., & Davies, D. R. (1962). Helix formation by guanylic acid. Proceedings of the National Academy of Sciences, 48(12), 2013–2018. https://doi.org/10.1073/pnas.48.12.2013
Henderson, E., Hardin, C. C., Walk, S. K., Tinoco, I., & Blackburn, E. H. (1987). Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell, 51(6), 899–908. https://doi.org/10.1016/0092-8674(87)90577-0
Neidle, S. (2017). Quadruplex nucleic acids as targets for anticancer therapeutics. Nature Reviews Chemistry, 1(5), 0041. https://doi.org/10.1038/s41570-017-0041
Neidle, S., & Balasubramanian, S. (Eds.). (2006). Quadruplex nucleic acids. Royal Society of Chemistry.
Oregon State University. (2015). Linus Pauling and the race for DNA: A documentary history. Retrieved 2 May, 2020, from http://scarc.library.oregonstate.edu/coll/pauling/dna/index.html
Pont, I., Martínez-Camarena, A., Galiana-Roselló, C., Tejero, R., Albelda, M. T., González-García, J., Vilar, R., & García-España, E. (2020). Development of polyamine-substituted triphenylamine ligands with high affinity and selectivity for G-quadruplex DNA. ChemBioChem, 21(8), 1167–1177. https://doi.org/10.1002/cbic.201900678
Watson, J. D., & Crick, F. H. C. (1953). Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature, 171, 737–738. https://doi.org/10.1038/171737a0