The Genetics of Human Migration
Tracing migrations through the genome

Various academic disciplines shed light on human migrations, helping us to reconstruct the past. Studying the genetic diversity of human populations today reveals past demographic and migratory events that have left an imprint on our genome. Armed with knowledge of migrations in prehistoric times, we can test hypotheses put forward in other scientific disciplines. Similarly, the distribution of genetic diversity in the future will largely depend on today’s extensive human migrations, facilitated by technological advances.
Keywords: genetic diversity, founder effect, genome, genetic gradient.
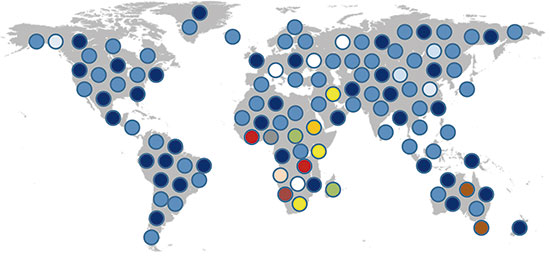
Figure 1. The figure shows genetic diversity in current human populations. The coloured circles highlight different genetic variants. Due to the African origins of mankind, Sub-Saharan populations have greater genetic diversity than other populations and greater effective population size. The genetic diversity observed outside Africa is a subset of African diversity due to the founder effect that took place some 50,000 years ago with the first great human migration. / David Comas
HUMAN ORIGINS AND THE TRAIL MIGRATION HAS LEFT IN OUR GENOME
Unlike the vast majority of living organisms, humans are a cosmopolitan species, scattered all over the planet and adapted to a wide variety of habitats. This widespread geographic distribution is the result of a series of migrations and mixtures of populations, which have taken place over a relatively short time in evolutionary terms. Various academic disciplines have reached a consensus on the recent African origin of our species: paleontological and archaeological data have located the oldest human sites in East Africa, and even linguistic studies based on phoneme diversity point to certain languages spoken by southern African hunter-gatherers as the most ancient. Furthermore, genetic data unquestionably support the fact that humans originated in Africa. So, how can we trace human origins through genetic data, and how can we track the subsequent migrations of human populations?
«Humans are a cosmopolitan species scattered all over the planet and adapted to a wide variety of habitats»
Our genome, comprised by DNA in the form of chromosomes held within the nuclei of our cells, carries the information to generate our bodily structures and functions. Replication and transmission of our genome are imperfect and so small errors, called random mutations or changes, are accumulated from generation to generation, leading to diversity between individuals. When these small changes occur in some specific regions of the genome they can lead to defects or dysfunction, but the vast majority of mutations are neutral, i.e., they do not cause any disorder, thus they accumulate in the germinal chromosomes and are transmitted to offspring. In this way, individuals, populations and the human species in general accumulate these changes in our genomes over time. This is the idea behind the so-called «molecular clock», mutations occur over time and thus genetic differences between groups of individuals, populations or species become greater as more time elapses from the point at which they separated.

On the American continent, we find lineages that originated in northeast Asia and dispersed throughout the New World from the Bering Strait, around 15,000 years ago. Currently Native American populations exhibit low diversity due to this relatively recent founder effect. The picture shows a Bolivian child. / Kris Krug
The study of these genetic variants of our genome reveals that current African populations have greater variation and therefore more genetic diversity than other human populations. What is more, much of the diversity in non-African populations is a subset of the variants found on the African continent. These findings support the Out-of-Africa theory on the first great migration of our species. This theory states that our species originated somewhere in Africa around 200,000 years ago and that after a period of diversification, with the consequent accumulation of mutations and diversity, some of these populations migrated out of the continent and colonised the rest of the planet. Comparing the genomes of African and non-African populations and using the molecular clock, we can estimate how much time has passed in terms of the differences accumulated, which reveal that the separation between these groups took place between 45,000 and 60,000 years ago. In other words, the first great human migration during which some individuals left the African continent took place in Palaeolithic times, when humans lived in small groups as hunter-gatherers (Henn et al. 2012).
«Genome replication and transmission are imperfect and cause small errors, called mutations, giving rise to diversity»
One of the main challenges facing the study of human population genetics is to discover the demographic processes experienced by African populations from their origin up until the first migration out of Africa. This is a very long time period, almost 150,000 years, during which African populations differentiated, accumulated genetic changes, migrated within the continent, and possibly mixed or – in some cases – even died out. We lack comprehensive genetic data for many African populations that would shed light on these questions. However, with the genetic data we do have, we can approximate the effective size of these African groups, i.e., how many individuals contributed to the diversity we observe today. Complete genome sequence data suggest the effective population size of our ancestors comprised between 12,000 and 15,000 individuals. This is the number of individuals required to generate and maintain the genetic diversity currently observed in these populations (Li and Durbin, 2011).
«Due to genetic drift, many variants are not randomly distributed in human populations but are clearly structured geographically»
On the other hand, the lower genetic diversity in populations outside Africa can be explained by the founder effect: a small group of humans carrying a subset of variants left the African continent and spread to the rest of the planet (Fig. 1). Measuring the genetic diversity of these non-African populations we can ascertain that, in the first great migration out of Africa, the migrant group harboured low genetic diversity, indicating that a small number of individuals left the continent. In fact, the group leaving Africa around 50,000 years ago did not exceed 1,000 or 2,500 individuals, yet their descendants colonised all the other regions of the world (Li and Durbin, 2011).

A classic example of the impact of human migration is the mixing of populations in the Americas after the arrival of Europeans and subsequent slave trade from Africa. In the photograph we can see two students with a teacher at a school in Lawrence (Massachusetts, USA). / Merrimack College
HUMAN COLONISATION OF THE PLANET
Humans are a species with low genetic diversity, due to our recent origin and the numerous founder effects over the years, which led to the loss of some of the original diversity each time we occupied new territories. However, this succession of founder effects as humans spread throughout the planet has led to geographically-structured genetic diversity, which enables us to reconstruct these human migrations. Genetic drift – i.e., the fact that some original genetic variants were lost by chance while others quickly became more frequent in populations – means that many genetic variants and their combination in the genome are not distributed randomly in human populations, but follow a clear geographic pattern. Many genetic variants are restricted to specific geographic areas, allowing us to draw a world map showing how these variants have spread. We call this phylogeography of genetic variants. The concept of this geographic structure involves studying the fragments of our genome that do not recombine, in other words, those which we inherit directly from our father (on the Y chromosome) or our mother (mitochondrial DNA) without genetic exchange between parents. In the last two decades, comprehensive studies of these uniparental genomes and their geographical distribution have enabled us to accurately track large continental colonisations and aspects of local migration. Some of the most interesting examples of the study of uniparental genomes include those on the colonisation of the American continent, which show how lineages originating in northeast Asia dispersed throughout the New World from the Bering Strait. This jump to the Americas about 15,000 years ago was accompanied by a strong founder effect, which dramatically reduced the genetic diversity of the first settlers of the Americas. Currently the Native American populations exhibit low diversity due to this relatively recent founder effect. Other examples of similar founder effects, though less dramatic than in the Americas, include the colonisation of Southeast Asia and the Pacific Islands or the expansion of populations from the Middle East to Europe. Indeed, we have been able to establish the major migratory routes undertaken by humans, in detail, and when these migrations occurred, by observing and quantifying the overall genetic diversity in current populations around the world, and by identifying specific variants of different geographical regions (Figure 2).
«The first great human migrations were in Palaeolithic times, when humans lived in small groups as hunter-gatherers»
Such human migrations have created gradients of genetic diversity, called genetic clines. As descendants of a population move and spread, generation after generation, they become genetically differentiated from the original population due to genetic drift. No abrupt changes or genetic breaks occur, but rather we can observe gradual changes in the frequencies of genetic variants in populations. We can follow the genetic footprints by studying those left in the path of these migrations. Furthermore, the genetic effect of these migrations may differ depending on whether dispersal takes place quickly – due to technological innovations – or passively into neighbouring territories because the gradients of genetic variants are distributed differently, geographically speaking, and leave a specific trail.
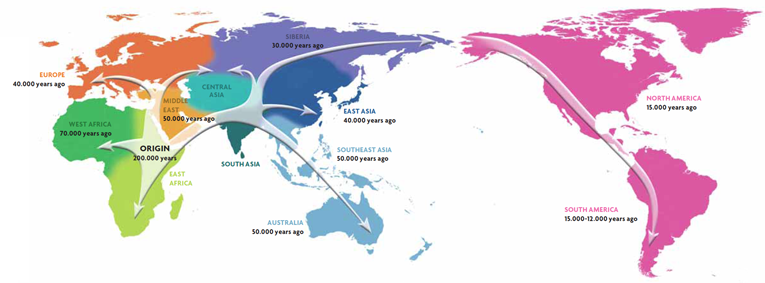
Figure 2. The major migratory routes followed by humans in prehistoric times. We have been able to determine great human migrations thanks to the degree of genetic diversity and incidence of specific genetic variants in particular geographical regions. / David Comas
MORE RECENT MIGRATIONS AND MIXED POPULATIONS
Once all the continents had been inhabited by humans, more recent demographic movements took place. Although we do not have data to establish the reason for migrations in many cases, we do know that climate changes or technological innovations prompted some of these demographic movements. In these cases, migrating populations did not enter territories unoccupied by humans and therefore these newcomers either replaced the resident population or mixed with it. This admixture of individuals from different populations has important genetic implications, which depend upon two factors: the proportion of newcomers who mix with the original population, and the genetic differences between the mixing populations. If a very small number of individuals migrated and mixed with another population comprising many individuals, genetic changes in the host population would be virtually unnoticeable, and would be undetectable by genetic analysis. Nor can we detect migratory effects when the mixing populations are genetically very similar, even though the number of newcomers is very high.
«Once all the continents had been inhabited by humans, more recent demographic movements took place, prompted by climate change or technological innovations»
A paradigmatic example of the impact of human migration can be found in the admixture of populations in the Americas, which took place after the arrival of Europeans and the subsequent slave trade of sub-Saharan Africans. Despite having a common origin, European, Native American and African populations had remained separate for tens of thousands of years and were, thus, genetically differentiated. Thus a large number of individuals from genetically distinct populations mixed, and it is relatively easy for us to detect the degree of admixture in the genomes of present Americans. We can even detect sex differences in the mix of individuals, as in the case of the Cuban population, where current uniparental genomes show that this admixture occurred mainly between European males and Amerindian and African women (Mendizabal et al., 2008), paying witness to European dominance and slavery. At the other extreme of genetic mixing, we can observe the impact of mixes involving a limited number of individuals and/or genetically similar populations. An example would be the Romanisation of the Iberian Peninsula, which probably involved a relatively small number of Romans compared to Iberians; furthermore, both populations had a recent common origin which would mean few genetic differences would have accumulated. Despite these limitations, the study of genetic markers and the analysis of complete genomes in human populations can help us to detect these small demographic impacts which were difficult to detect before.
«Current human migrations are extensive, and large-scale population mixing is favouring the homogenisation of human genetic diversity»
Diasporas represent a special case within human migrations. This happens when a whole group of individuals migrates from its place of origin to a distant place without leaving individuals behind along the way, and without mixing with the inhabitants of the territory where they settle. This is the case of Jews or Gypsies, whose populations conserved genetic variants of the Middle East or India, respectively, despite being surrounded by European populations. These populations have experienced inbreeding and isolation processes, and have thus maintained the original genetic variants that were present prior to the diaspora (Behar et al., 2010; Mendizabal et al., 2012).

We can detect sex differences in the mix of individuals, as in the case of the Cuban population, where current uniparental genomes show that this admixture occurred mainly between European males and Ameridian and African women, paying witness to European dominance and slavery. The image shows several children playing football on the streets of Camagüey (Cuba). / Gerry Balding
Genetic studies of current populations can help us to establish the migratory movements of humans in prehistoric times, and allow us to test the hypotheses proposed by other scientific disciplines. Technological advances in recent centuries have increased and speeded up current human migrations, and large-scale population mixing is favouring the homogenisation of human genetic diversity and reducing genetic differences between populations, forged over millennia. However, unlike the vast majority of living organisms, humans have a complex cultural and social diversity which has sometimes acted as a genetic barrier or as a factor enhancing population mixing and migrations. Future distribution of genetic diversity will largely depend upon these social factors that favour or hinder human migrations.
On migrations, founder effects and mixed populationsThe history of the Roma |
 European Roma originated in the northwest of India and a few individuals left the region around 1,500 years ago. The diaspora of this small group led to a drastic reduction in genetic diversity. The photograph shows a nomadic Gypsy community in Maharashtra (India). / Poonam Agarwal The Roma people, also known as Gypsies, are one of the most interesting populations – genetically speaking – on the European continent, due to their demographic history. European Roma currently number around ten million people, scattered in disperse groups across the continent, but most reside in the Balkans and the Iberian Peninsula. Although cultural traits and genetic backgrounds do not always coincide, linguistics and physical anthropology suggest that the Roma originated in the Indian subcontinent. Thus, genetic data have enabled us to hone in on the history of the Roma people. In the last decade, studies of Gypsy groups’ uniparental genomes (Y-chromosome and mitochondrial DNA) reveal a mixture of European lineages and lineages found only in the Indian subcontinent, which supports observations based on linguistic and anthropological data. Moreover, the diversity of these lineages (European and Indian) is quite low, which suggests that current Roma come from a few individuals of Indian origin with European genetic introgression. However, uniparental lineages represent only a small fraction of our entire genome and so to hone in on the demographic history of the Roma, we must analyse thousands of variants of the genome of numerous Roma groups. In December 2012, in the journal Current Biology (Mendizabal et al., 2012) we reported the results of analysing nearly one million genetic variants in different individuals of various Roma groups and we compared them with other European, Indian and Middle Eastern populations. Analysis of their genetic diversity revealed that European Roma originated in the northwest of India and a few individuals left the region some 1,500 years ago. The diaspora of this small group led to a drastic reduction in genetic diversity (50 % of that observed in India) due to a strong founder effect. The ancestors of the Gypsies migrated quickly through the Middle East, with very little genetic admixture occurring with populations encountered on the way, and eventually arrived in Europe and settled in the Balkans. Genetic data suggest that this initial population fragmented into smaller groups (creating further founder effects), which began to scatter throughout Europe around 900 years ago. During this dispersal, the Roma’s ancestors from India mixed with various European groups and due to this admixture today’s Gypsies have genetic variants typical of India and Europe. Nonetheless, this genetic mixing is not uniform in all Roma groups. Our analyses show that Roma populations in the Balkans have fewer typically European variants than Roma populations in the Iberian Peninsula, which shows that after dispersing through Europe, the Iberian Gypsies mixed more with neighbouring populations than the Balkan Roma did. We were even able to establish that in the Balkan groups the admixture with non-Roma populations occurred fairly recently, as the European chromosomal fragments found in these Gypsy groups are unusually long. These long fragments indicate that chromosomal recombination (a genomic process whereby paired chromosomes intersect) has not had time to occur on a large scale. In short, genetic data have shed light on some aspects of the demographic history of the social relations between Roma groups, supporting hypotheses posed by other disciplines such as linguistics and anthropology. |
Behar, D. M. et al., 2010. «The Genome-Wide Structure of the Jewish People». Nature, 466: 238-242. DOI: <10.1038/nature09103>.
Henn, B. M.; Cavalli-Sforza, L. L. and M. W. Feldman, 2012. «The Great Human Expansion». Proceedings of the National Academy of Sciences USA, 109: 17758-17764. DOI: <10.1073/pnas.1212380109>.
Li, H. and R. Durbin, 2011. «Inference of Human Population History from Individual Whole-Genome Sequences». Nature, 475: 493-496. DOI: <10.1038/nature10231>.
Mendizabal, I. et al., 2008. «Genetic Origin, Admixture, and Asymmetry in Maternal and Paternal Human Lineages in Cuba». BMC Evolutionary Biology, 8: 213. DOI: <10.1186/1471-2148-8-213>.
Mendizabal, I. et al., 2012. «Reconstructing Population History of European Romani from Genome-Wide Data». Current Biology, 22(24): 2342-2349. DOI: <10.1016/j.cub.2012.10.039>.