The environmental watchdogs
Wildlife as sentinels of antimicrobial resistance pollution in the environment in Catalonia

The increasing prevalence of antimicrobial resistance (AMR) in both humans and livestock is attributed largely to the overuse and misuse of antimicrobials. The alarming emergence of this resistance in human and veterinary medicine has activated awareness for monitoring the levels of AMR pollution in the environment. In this report, the emergence of genes conferring resistance to human last-resort antibiotics is described in a wide diversity of wild animals. It suggests that wildlife can be good sentinels of AMR environmental pollution, especially in highly populated areas. Moreover, wild animals can also contribute in the dissemination of AMR bacteria and genes in the environment and represent a zoonotic risk for the population who are exposed to them.
Keywords: antimicrobial resistance, beta-lactamases resistance genes, Catalonia, colistin-resistance genes, wildlife.
In the last decades, the prevalence of opportunistic and antimicrobial resistant bacteria (AMRB) associated with nosocomial infections has increased considerably in hospital settings. The overuse and misuse of antibiotics in human and veterinary medicine have led to the spread of antimicrobial resistance (AMR) pathogens, becoming a global health problem (Pitout & Laupland, 2008). The alarming emergence of AMR in human and veterinary medicine has activated awareness for monitoring the levels of this kind of resistance pollution in the environment. Furthermore, the implementation of control measures under the One Health approach are urgently needed to reduce the impact of anthropogenic pressure in the environment. Wildlife has been suggested to be a good sentinel of environmental health because of its close proximity with human populations, domestic animals, and natural ecosystems. Moreover, wildlife can act as long-term asymptomatic carriers of zoonotic pathogens (Tsiodras et al., 2008), including viruses (e.g., influenza and West Nile viruses), bacteria (e.g., Salmonella spp., Campylobacter spp., methicillin resistant Staphylococcus aureus), or parasites (e.g., Cryptosporidium spp.). Accordingly, they can contribute to the dissemination of these pathogens to livestock and to urban areas, and thereby increasing the risk of infection for animals and humans.
Extended-spectrum β-lactamases (ESBLs) and AmpC-type β-lactamases (ampC) are the most common enzymes that confer resistance to broad-spectrum antibiotic cephalosporins among members of the family Enterobacterales. These β-lactamases (bla) have extensively diversified in response to the clinical use of new generation drugs: cephalosporins, carbapenems, and monobactams. β-lactamases enzymes can be classified in three groups. The first group contains cephalosporinases encoded in the chromosome of many Enterobacterales, such as ampC, blaCMY, blaACT, blaFOX, and blaMIR. Some variants of these enzymes have also been detected in plasmids. The second group, serine beta-lactamase, represents the largest group with a broad spectrum against penicillins, cephalosporins, and carbapenems. They include the blaTEM, blaSHV, blaCTX, blaOXA, and blaKPC enzymes. These enzymes are mostly encoded by genes located in plasmids that can be horizontally transferred to different bacteria genera. Finally, the third group, metallo-beta-lactamases, are zinc dependent and include blaNDM, blaIMP, blaVIM, and blaSPM enzymes. The emergence of resistance to carbapenems – last resort drugs used in hospital settings for the treatment of severe infections caused by β-lactam resistant Enterobacteria – has compromised the therapeutic options in human health (Tzouvelekis et al., 2012). Specially, the OXA-48 variant of carbapenemases is becoming highly prevalent in human clinical infections in Europe (Grundmann et al., 2017).
Other human last resort antibiotic is the colistin. Mobile colistin resistance (mcr) genes are plasmid-mediated resistance genes recently discovered in humans and food animals (Skov & Monnet, 2016). The mcr genes are known to confer resistance against colistin sulphate – one of the last effective drugs for the treatment of multi drug resistence (MDR) gram-negative bacteria infections (Sun et al., 2018). Colistin has been used in veterinary medicine during the last decades for the treatment of gastrointestinal infections in livestock, principally in pigs and poultry (EMA, 2016) but also as preventive mass-medication against colibacillosis in piglets. Consequently, livestock is considered the main reservoir of mcr gene selection and dissemination worldwide. However, nowadays there is a worldwide trend to limit colistin usage in animal production due to significant public health concern related to emergence and transmission of resistance genes (Sun et al., 2018).
Antimicrobial resistance studies in wildlife in Catalonia
The dissemination of extended-spectrum β-lactamases has been studied widely in Enterobacterales from humans and livestock, whereas studies concerning the environment, including wildlife, are still lacking. Catalonia, an autonomous community located in the northeast of Spain, has an estimated population over 7.8 million with a density of 242.3 inhabitants per km2. Catalonia is a region with a high density of livestock farms, mainly pig farms, leading the Spanish pig production with another record level of 32.8 million swine animals in 2020, a 5 % increase compared to the previous year, according to Eurostat data.
Under these conditions, wild animals attended at the Wildlife Rehabilitation Centre of Torreferrussa (located at the Barcelona county) were the subject of different epidemiological studies. All animals were examined and tested using cloacal or rectal swabs on arrival at the centre before receiving any pharmacologic or antimicrobial treatment. The most frequent cause of hospitalization was related to anthropogenic origin due to direct persecution (gunshot, poisoning, illegal captivity or traps) or to involuntary human induced threats (collisions with vehicles, fences or electric lines and electrocution). Sampling methods and handling protocols of animals were in agreement with the Catalan Wildlife Service who stipulates the management protocols and ethical principles according to the Spanish legislation.
Wild birds
The first studies conducted revealed a significant prevalence of antimicrobial-resistant bacteria in wild raptors without any record of antimicrobial treatment. In particular, 10 % and 7.4 % of wild raptors were positive to multi-drug resistence Salmonella and quinolone-resistant Campylobacter strains respectively (Molina-López et al., 2011). Among the large diversity of Salmonella serovars found in that study, the ampicillin, chloramphenicol, streptomycin, sulphonamide and tetracycline (ACSSuT) multiresistant Salmonella Typhimurium DT104, was identified in common buzzards (Buteo buteo) and a little owl (Athene noctua).
In another study, 6.3 % of raptors (Bubo bubo, Tyot alba, Strix aluco, Falco tinnunculus, Gyps fulvus) were also positive for Salmonella with the predominant detection of S. Typhimurium monophasic 4,12:i:- variant with a ampicillin, streptomycin, sulfonamides, and the tetracycline (ASSuT) resistant profile expanded to colistin and chloramphenicol in some isolates (Molina-López et al., 2015). In Europe, monophasic strains are often characterized by resistant ASSuT profile, and there is evidence that wild boars (Sus scrofa) are probably one of the reservoirs of strains of that phage type in Spain (Mateu et al., 2002) and Catalonia (Mejía et al., 2006).
On the other hand, the frequency of fluoroquinolone resistant Campylobacter isolates seems consistently responsive to local antimicrobial selection pressure. The presence of quinolone-resistant isolates in the other wild birds could be due to the ingestion of prey. Moreover, in species with scavenging food habits, such as the griffon vulture (Gyps fulvus), direct ingestion of antibiotic residues and antibiotic resistant bacteria from livestock carrion or infected carcasses of wildlife could be a source of infection.
The implementation of genotyping analyses allowed to take a step forward with the AMR studies in wildlife. Thus, in 2019 we reported, for the first time in Spain, the presence of cephalosporin resistance encoding genes in a large variety of Enterobacterales in wild mammals – principally in hedgehogs and mustelids – and wild birds – mainly in raptors – (Darwich et al., 2019). Principally, the overall prevalence of AMRB in that study was 13 % (17.3 % in wild mammals and 11.5 % in wild birds). Hedgehogs showed the highest prevalence (13.5 %) within the mammal specimens, and raptors within the avian specimens (7.3 %). Most of the AMRB isolated from this wildlife contained a high diversity of resistant genes (50 % blaCMY-2, 23 % blaSHV-12, 20 % blaCMY-1 y 18 % blaCTX-M-15) that were frequently found in nosocomial infections in human hospitals, such as Escherichia coli, Klebsiella pneumoniae, Citrobacter freundii, Enterobacter cloacae, Serratia marcescens and Proteus mirabilis.
Hedgehogs
The European hedgehog (Erinaceus europaeus) is a common and widely distributed wild species in Europe. Hedgehogs are small, nocturnal, spiny-coated insectivores that live in a variety of habitats, generally in close contact with humans, but also with livestock in the countryside. In recent years, hedgehog populations increasingly inhabit areas with human activity, such as gardens in residential areas or green areas in big cities. Some of the consequences of this close hedgehog-human interaction is that people usually feed these animals and thus increases the risk of human physical contact. Thus, we conducted a study with a larger cohort of European hedgehogs from the same area of Catalonia, showing that 36.8 % of the animals were carriers of AMR genes, with a special occurrence of human nosocomial bacteria harbouring ESBL and AmpC genes (Garcias et al., 2021). Moreover, K. pneumoniae was the bacteria with the highest proportion of resistance genes, followed by E. coli and C. freundii. The most frequently detected genes were also blaCTX-M-15 (19.3 %), blaSHV-28 (10.5 %), blaCMY-1 (9.7 %), and blaCMY-2 (8.8 %). It is interesting to remark that more than half of the enterobacteria presented MDR phenotype, with an extended profile (XDR) in 30 % of the isolates. Furthermore, we have described the presence of carbapemenase OXA-48 variant genes in Enterobacterales from hedgehogs inhabiting highly populated urban areas of Catalonia (Darwich et al., 2019; Garcias et al., 2021). As carbapenems are not allowed in food animals, the increasing presence of carbapenemases genes in livestock, companion animals, and wild animals is alarming evidence of the anthropogenic environmental contamination.

Hedgehogs are small, nocturnal, spiny-coated insectivores that live in a variety of habitats. In recent years, hedgehog populations increasingly inhabit areas with human activity, such as gardens in residential areas or green areas in big cities. Some of the consequences of this close hedgehog-human interaction is that people usually feed these animals and thus increases the risk of human physical contact. / Photo: Michale Gäbler – Wikipedia
One possible explanation for the highest prevalence of β-lactamase resistance genes detected in hedgehogs could be found in the feeding habits and the location of these animals. It must be related to exposure to sewage water or soils contaminated with antimicrobial resistant bacteria and antimicrobial residues. Earthworms, a key biological component of soil, have been shown to contribute to spreading antimicrobial resistance, as they can acquire antibiotics directly from the soil. Thus, considering that most of the hedgehog’s diet is based on earthworms and other invertebrates, hedgehogs could be more directly exposed to these antibiotic substances and resistance genes than other animal species (Garcias et al., 2021).
Wild boars
Following the same research objective, wild boars (n = 200) foraging in metropolitan area of Barcelona were examined for the presence of E. coli carrying critical ESBL antimicrobial resistance genes (ARG) and other zoonotic pathogens (Darwich et al., 2021). Wild boar (Sus scrofa) populations are rising steadily worldwide, which is an increasing cause of concern since the wild boar has been identified as a reservoir of zoonoses and food-borne pathogens. On the other hand, the wild boar from the Metropolitan Area of Barcelona is a suitable sentinel to monitor the impact of the use of anthropogenic food resources. The results of this study showed 8 % of animal carriers of β-lactamases resistance genes: blaCMY-2 (3.0 %), blaTEM-1b (2.5 %), blaCTX-M-14 (1.0 %), blaSHV-28 (1.0 %), blaCTX-M-15 (0.5 %) and blaCMY-1 (0.5 %). Moreover, toxigenic Clostridioides difficile TcdA+ was detected in two wild boars, which was the first report of this pathogen in wild boars in Spain. These results indicate the value of this game species as a sentinel of antimicrobial-resistant bacteria (AMRB) and zoonotic agents in urban environments. Moreover, the wild boars foraging in urban and peri-urban locations were more exposed to AMRB sources than the wild boars dwelling in natural environments. Concretely, β-lactam resistance genes were more frequently isolated from the urban and peri-urban wild boars (14 % of prevalence) than in the forest ones (2 % of prevalence) (Darwich et al., 2021).
Semi-aquatic-life animals
Studies conducted in wild birds and mammalian species showed that these animals can serve as a reservoirs and potential vectors for the spread of resistant bacteria and antimicrobial resistance genes (Darwich et al., 2019). However, there are limited data regarding the role of semi-aquatic animals as sentinel of environmental health. For this reason, the most recent study conducted by our team was focussed on the occurrence of AMRB and ARG in wild semi-aquatic animals from highly populated urban areas and intensive farming regions of Catalonia, principally pork and poultry production (Mengistu et al., 2022).
Principally, cloacal and rectal swab samples were collected from invasive species such as Pond sliders (Trachemys scripta) and American minks (Neovison vison). Accordingly, 55.2 % isolates were identified as AMRB. E. coli and Pseudomonas fluorescens were among the bacteria most frequently isolated in all animal species, but other nosocomial agents such as K. pneumonia, Salmonella spp, or C. freundii were also prevalent. The phenotypic susceptibility testing showed the highest resistance in macrolides and β-lactams. In addition, the presence of ESBL/Ampc genes were found in 15.4 % of turtles and 8.4 % of American minks. The genotyping frequency was tetM (20.6 %), blaCMY-2 (13 %), ermB (6.1 %), blaCMY-1 (4.6 %), blaCTX-M-15 (3.1 %) and mcr-4 (0.8 %). Turtles had a larger prevalence of MDR bacteria and AMR genes than mustelids, but mcr-4 colistin resistance gene was detected in American minks. Furthermore, the distribution of β-lactam, erythromycin, tetracycline and colistin-resistance genes were concentrated in the metropolitan areas of big cities and in regions with a high pig farm production. Specifically, the highest frequency of animals positive to ESBL/ampC genes were located in populated areas comprising big metropolitan regions (big human hospitals) and the highest frequency of animals carrying tetracycline and erythromycin (tetM/emrB) genes or mcr-4 colistin-resistance gene were from areas with a high intensive livestock production (Mengistu et al., 2022).
In general, mcr genes, principally mcr-1 has been reported worldwide in Enterobacterales isolated from humans, livestock, companion animals, food and wildlife (Skov et al., 2016). Moreover, mcr-1 and mcr-4 are the most frequent colistin-resistance genes reported in Enterobacterales in pigs from Spain (Aguirre et al., 2020; Vidal et al., 2020). Thus, the presence of mcr-4 in a K. pneumoniae from an American mink habiting one of the counties with the highest pig density is a coherent result. The previous abuse of colistin sulphate in pig production as a preventive for controlling post-weaning diarrhoeas could be the cause of a selective pressure for colistin resistant bacteria. In fact, from 2013 to 2015, Spain was the European country with the highest sales of polymyxins for food-producing animals (EMA, 2016), and with a significant prevalence of colistin-resistance genes, principally mcr-4 (Aguirre et al., 2020). The Spanish Agency for Medicines and Health Products established a drastic reduction (97 %) in the use of colistin in swine from 2015 to 2018 in Spain, with a positive impact in the decrease in colistin resistance genes (Aguirre et al., 2020). However, the presence of these colistin resistant genes in natural waters can suggest that the reduction in the use of this antimicrobial is not enough to prevent gene selection in live pigs or that river contamination come from stable mcr genes in old non-treated purines. In conclusion, semi-aquatic wild animals are good sentinel of environmental (water) pollution with resistant bacteria and antimicrobial resistance genes.
A major cause of concern
Most of the extended-spectrum β-lactamases genes detected in wildlife from Catalonia are also found in hospital settings frequently, suggesting a relationship between bacteria from wildlife and health-care facilities (Figure 1). Besides, this high prevalence of human nosocomial-like bacteria with β-lactamase resistance genes in the wildlife populations of urban surroundings in Catalonia is a major cause of concern, because these animals have never been treated with an antibiotic; thus, the presence of ARG must be related to exposure to sewage water or soils contaminated with AMRB or antimicrobial residues. Sewage treatment plants are not completely efficient at removing all antibiotic residues and AMRB (Karkman et al., 2018), and contaminated water is spread into rivers and seas. Moreover, livestock manure is applied to land and can also contain it (Han et al., 2021), enriching the resistome in both soils and waters (Zhou et al., 2020).
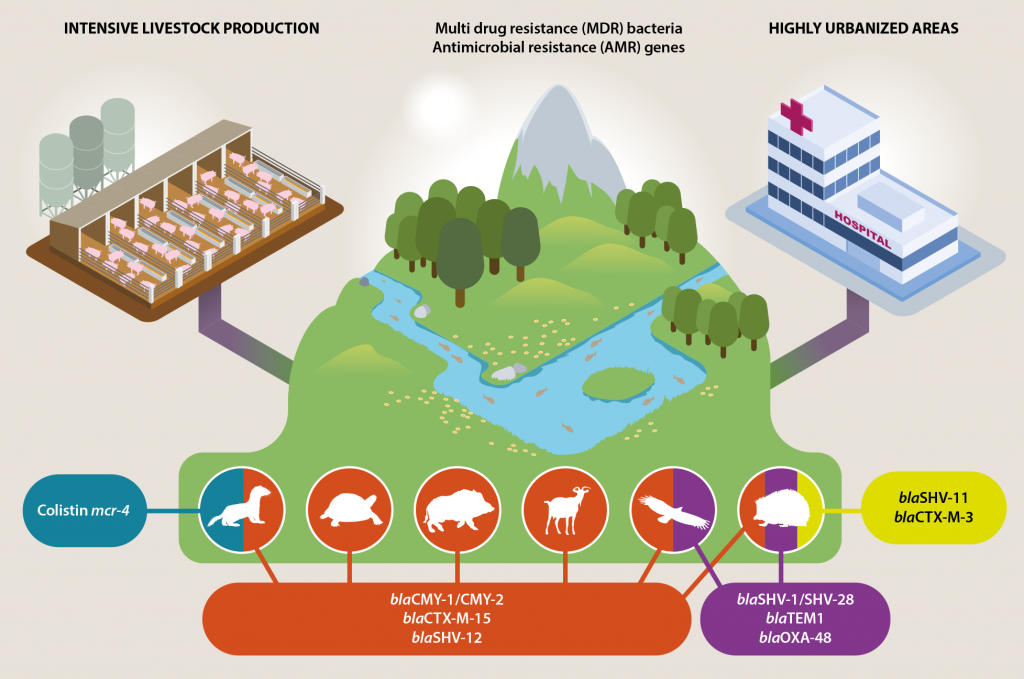
Figure 1. Graphic summary of antimicrobial resistance genes detected in the wildlife (provided by L. Darwich). Waste from facilities such as intensive livestock production farms and hospitals contributes to polluting the environment with resistant bacteria and resistance genes to many antibiotics that are critical for human and veterinary medicine. Wild animals in close proximity to these areas carry these antibiotic resistant genes and bacteria. Therefore, it is important to keep an eye on them to know the degree of environmental contamination that exists in a specific area and also because they can disseminate these bacteria and resistance genes to other territories. / Source: L. Darwich / Illustration: Xavier Sepúlveda
The relationship between levels of AMR in the environment and the occurrence in humans is complex: there are numerous combinations of antimicrobials, bacterial strains, mobile genetic elements (MGEs) and wildlife species, each with their own dynamics. A recent study (Wee et al., 2020) focused on the relationship between AMRB in humans and livestock have reported that horizontal gene transfer of ARG via MGEs such as plasmids, expands the human–livestock interface beyond the confines of clonal transmission. As well, genomic investigations have revealed highly related ARG and mobile elements in bacterial lineages across humans and livestock.
The application of high-resolution genome sequencing has the potential to be a powerful tool to help with our baseline understanding of the pathways of AMRB dissemination, or their resistance determinants from human to wildlife and vice versa. This will be key to the development of effective policies on antimicrobial stewardship and infection control for human-animal-environmental health.
One Health approach is urgently needed
In conclusion, asymptomatic wildlife without any record of antimicrobial treatment may carry AMRB resistant to drugs used in human or veterinary medicine, and therefore act as a reservoirs and sentinels of such resistant bacteria. Furthermore, wildlife can represent a zoonotic risk for staff of Wildlife Rehabilitation Centres and the general population in close and continuous contact with wild animals. In consequence, it is crucial to raise awareness about the strong interconnection between habitats and compartments induced by multiple exchange routes, which implies that AMR issues must be tackled under the One Health approach. Therefore, One Health approach is urgently needed in highly populated regions and with intensive livestock production like Catalonia.
References
Aguirre, L., Vidal, A., Seminati, C., Tello, M., Redondo, N., Darwich, L., & Martín, M. (2020). Antimicrobial resistance profile and prevalence of extended-spectrum beta-lactamases (ESBL), AmpC beta-lactamases and colistin resistance (mcr) genes in Escherichia coli from swine between 1999 and 2018. Porcine Health Management, 6, 4–9. https://doi.org/10.1186/s40813-020-00146-2
Darwich, L., Seminati, C., López-Olvera, J. R., Vidal, A., Aguirre, L., Cerdá, M., Garcias, B., Valldeperes. M., Castillo-Contreras, R., Migura-Garcia L., Conejero, C., & Mentaberre, G. (2021). Detection of beta-lactam-resistant Escherichia coli and toxigenic Clostridioides difficile strains in wild boars foraging in an anthropization gradient. Animals, 11, 1585. https://doi.org/10.3390/ani11061585
Darwich, L., Vidal, A., Seminati, C., Albamonte, A., Casado, A., Lopez, F., Molina-López, R. A., & Migura-Garcia, L. (2019). High prevalence and diversity of extended-spectrum β- lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLOS One, 14(8), e0210686. https://doi.org/10.1371/journal.pone.0210686
EMA. (2016). Updated advice on the use of colistin products in animals within the European Union: Development of resistance and possible impact on human and animal health. http://www.ema.europa.eu/docs/en_GB/document_library/Press_release/2016/07/WC500211081.pdf
Garcias, B., Aguirre, L., Seminati, C., Reyes, N., Allepuz, A., Obón, E., Molina-Lopez, R. A., & Darwich, L. (2021). Extended-spectrum b-lactam resistant Klebsiella pneumoniae and Escherichia coli in wild European hedgehogs (Erinaceus europeus) living in populated areas. Animals, 11, 2837. https://doi.org/10.3390/ani11102837
Grundmann, H., Glasner, C., Albiger, B., Aanensen, D. M., Tomlinson, C. T., Andrasević, A. T., Cantón, R., Carmeli, Y., Friedrich, A. W., Giske, C. G., Glupczynski, Y., Gniadkowski, M., Livermore, D. M., Nordmann, P., Poirel, L., Rossolini, G. M., Seifert, H., Vatopoulos, A., Walsh, T., … EuSCAPE working group. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): A prospective, multinational study. The Lancet Infectious Diseases, 17, 153–163. https://doi.org/10.1016/S1473-3099(16)30257-2
Han, B., Yang, F., Tian, X., Mu, M., & Zhang, K. (2021). Tracking antibiotic resistance gene transfer at all seasons from swine waste to receiving environments. Ecotoxicology and Environmental Safety, 219, 112335. https://doi.org/10.1016/j.ecoenv.2021.112335
Karkman, A., Do, T. T., Walsh, F., & Virta, M. P. J. (2018). Antibiotic-resistance genes in waste water. Trends in Microbiology, 26(3), 220–228. https://doi.org/10.1016/j.tim.2017.09.005
Mateu, E. M., Martin, M., Darwich, L., Mejia, W., Frias, N., & Garcia Peña, F. J. (2002). Antimicrobial susceptibility of Salmonella strains isolated from swine in Catalonia, Spain. The Veterinary Record, 150, 147–150. https://doi.org/10.1136/vr.150.5.147
Mejía, W., Casal, J., Zapata, D., Sánchez, G. J., Martín, M., & Mateu, E. (2006). Epidemiology of Salmonella infections in pig units and antimicrobial susceptibility profiles of the strains of Salmonella species isolated. The Veterinary Record, 159, 271–276. https://doi.org/10.1136/vr.159.9.271
Mengistu, T. S., Garcias, B., Castellanos, G., Seminati, C., Molina-Lopez, R. A., & Darwich, L. (2022). Occurrence of multidrug resistant Gram-negative bacteria and resistance genes in semi-aquatic wildlife–Trachemys scripta, Neovison vison and Lutra lutra–as sentinels of environmental health. Science of the Total Environment, 830, 154814. https://doi.org/10.1016/j.scitotenv.2022.154814
Molina-López, R. A., Valverdú, N., Martin, M., Mateu, E., Obon, E., Cerdà-Cuéllar, M., & Darwich, L. (2011). Wild raptors as carriers of antimicrobial-resistant Salmonella and Campylobacter strains. The Veterinary Record, 168(21), 565. https://doi.org/10.1136/vr.c7123
Molina-López, R. A., Vidal, A., Obón, E., Martín, M., & Darwich, L. (2015). Multidrug-resistant Salmonella enterica serovar Typhimurium monophasic variant 4,12:i:- isolated from asymptomatic wildlife in a Catalonian wildlife rehabilitation center, Spain. Journal of Wildlife Diseases, 51(3), 759–763. https://doi.org/10.7589/2015-01-019
Pitout, J. D., & Laupland, K. B. (2008). Extended-spectrum beta-lactamase-producing Enterobacteriaceae: An emerging public-health concern. The Lancet Infectious Diseases, 8(3), 159–66. https://doi.org/10.1016/S1473-3099(08)70041-0
Skov, R. L., & Monnet, D. L. (2016). Plasmid-mediated colistin resistance (mcr-1 gene): Three months later, the story unfolds. Eurosurveillance, 21, 30155. https://doi.org/10.2807/1560-7917.ES.2016.21.9.30155
Sun, J., Zeng, X., Li, X., Liao, X., Liu, Y., & Lin, J. (2018). Plasmid-mediated colistin resistance in animals: Current status and future directions. Animal Health Research Reviews, 18, 136–152. https://doi.org/10.1017/S1466252317000111
Tsiodras, S., Kelesidis, T., Kelesidis, I., Bauchinger, U., & Falagas, M. E. (2008). Human infections associated with wild birds. The Journal of Infection, 56, 83–98. https://doi.org/10.1016/j.jinf.2007.11.001
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., & Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clinical Microbiology Reviews, 25, 682–707. https://doi.org/10.1128/CMR.05035-11
Vidal, A., Aguirre, L., Seminati, C., Tello, M., Redondo, N., Martín, M., & Darwich, L. (2020). Antimicrobial resistance profiles and characterization of Escherichia coli strains from cases of neonatal diarrhea in Spanish pig farms. Veterinary Sciences, 7(2), 48. https://doi.org/10.3390/vetsci7020048
Wee, B. A., Muloi, D. M., & van Bunnik, B. (2020). Quantifying the transmission of antimicrobial resistance at the human and livestock interface with genomics. Clinical Microbiology and Infection, 26(12), 1612–1616. https://doi.org/10.1016/j.cmi.2020.09.019
Zhou, S., Zhu, D., Giles, M., Daniell, T., Neilson, R., & Yang, X. R. (2020). Does reduced usage of antibiotics in livestock production mitigate the spread of antibiotic resistance in soil, earthworm guts, and the phyllosphere? Environment International, 136, 105359. https://doi.org/10.1016/j.envint.2019.105359