Dodging magic bullets
The evolution of bacterial antibiotic resistance
Antibiotic-resistant bacteria currently represent one of the main public health problems and recent predictions indicate that they will soon become the world’s leading cause of death.
The ill-fated journey from the introduction of antibiotics into clinical practice to the current threat of a post-antibiotic era has run its course in just a few decades. Thus, the evolution of antibiotic resistance is probably the most spectacular example of evolution of a biological system innovation that we have had the opportunity to observe in real time. This text discusses some of the evolutionary and molecular keys that have allowed bacteria to go down this path.
Keywords: evolutionary innovation, antibiotic resistance, bacteria, plasmids, horizontal gene transfer.
Introduction
Living beings’ potential to exploit new ecological opportunities depends on their ability to innovate. The evolution of innovations consists in the acquisition of new characters that allow organisms to expand their niche and face new environmental challenges. Some classic examples of evolutionary innovation are the appearance of flowers in plants or, in animals, the ability to fly (Mayr, 1963). In many cases, these innovations are self-evident at the phenotypic level, and may even be spectacularly obvious; we just need to look up and see a flock of birds crossing the sky to see such examples. However, the molecular bases underlying these adaptations are not so apparent. How are innovations generated? What are the genetic foundations of this process? These questions have been the subject of study for decades and remain unsolved despite everything we have learned about them (for more information about the origin of innovations, see Wagner, 2015). In this article we will try to illustrate some of the keys to the process of evolutionary innovation through one of the most dramatic current examples: the evolution of bacterial antibiotic resistance.
«700,000 people die each year globally as a result of antibiotic-resistant infections»
An ignored warning
Antibiotics are undoubtedly one of the most important discoveries in human history. These «magic bullets», as Paul Ehrlich defined them as early as the twentieth century (Tan & Grimes, 2010), are molecules capable of specifically inhibiting bacterial growth and have been the basis for the development of modern medicine and surgery. They do indeed seem to be magic bullets; antibiotics are incredibly efficient at treating bacterial infections. Paradoxically, this great effectiveness has also been the reason for their decline, because this precious resource has been excessively and often inappropriately used for decades, which has allowed bacteria to evolve antibiotic resistance. This phenomenon should not surprise us, because in his 1945 Nobel Prize acceptance speech, Alexander Fleming had already warned us of the ability of microorganisms to develop antibiotic resistance and the danger that this would represent (Fleming, 1945). Unfortunately, the warning was ignored, and antibiotics were abused from the time they were introduced into clinical practice in the mid-twentieth century. This led to a constant increase in the frequency of resistance by bacterial pathogens. In fact, the spread of antibiotic resistance among pathogens of clinical interest has now become one of our main public health problems (Figure 1) and is recognised by every international health organisation, the United Nations, and even the International Monetary Fund (CDC, 2013; ECDC, 2013; Jonas, Parry, Chisholm, Banatvala, & Ladminarayan, 2014; WHO, 2016). The World Health Organisation, for instance, stresses that «without urgent action, we are heading for a post-antibiotic era, in which common infections and minor injuries can once again kill» (WHO, 2018). According to recent estimates, approximately 700,000 people die each year globally as a result of antibiotic-resistant infections (O’Neill, 2016). These predictions also indicate that, if the current trend of antibiotic resistance spread is not reversed, by 2050 resistant infections could become the leading cause of death in the world, potentially leading to ten million deaths annually.
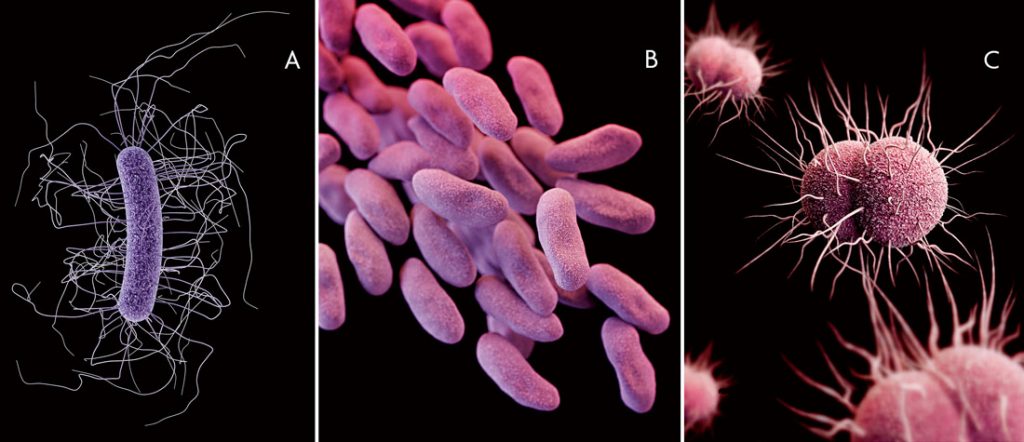
Figure 1. In 2013, the Center for Disease Control and Prevention (CDC) released a report cataloguing the main threats to public health caused by antibiotic-resistant pathogens of clinical interest (CDC, 2013). The picture shows 3D renderings of the structure of three bacteria classified as «urgent threats» in the report (from left to right): A) Clostridium difficile (which causes life-threatening diarrhoea and colitis); B) Clones pertaining to the Enterobacteriaceae family, resistant to carbapenem antibiotics, and C) Neisseria gonorrhoeae antibiotic-resistant strains (the bacterium responsible for gonorrhoea). / CDC / James Archer
Adapting to antibiotics
The evolution of bacterial antibiotic resistance is a simple and elegant example of adaptation by natural selection (Figure 2). A bacterium acquires a resistance mechanism and it is selected due to the presence of the antibiotic. In the absence of the antibiotic, the resistance mechanism often imposes a biological cost in terms of reducing the bacterium’s fitness or biological efficacy (defined as a decrease in its reproductive rate). However, this reduction in biological efficacy can be compensated for by mutations that alleviate the cost associated with antibiotic resistance. Taking this dynamic into account, it is easy to understand how overusing antibiotics has promoted the global spread of antibiotic resistance.
«By 2050, resistant infections could become the leading cause of death in the world, potentially leading to ten million deaths annually»
Antibiotic resistance is therefore a fascinating example of evolutionay innovation, because an antibiotic-sensitive bacteria needs to innovate to acquire resistance. What is even more fascinating is the fact that to study this evolutionary innovation we do not need to resort to fossil remains, rather, we have witnessed it in real time over the last few decades. Moreover, we can even partially reproduce this process of evolution in our laboratories via a process known as «experimental evolution» (Buckling, Craig Maclean, Brockhurst, & Colegrave, 2009). Finally, and this is a crucial point, thanks to technological advances, today we can exactly determine the genetic bases underlying the evolution of antibiotic resistance in bacteria. In short, all of the above puts us in an ideal position to answer some of the key questions about the evolution of innovations responsible for antibiotic resistance, which we will do in the following lines.
Routes to resistance
The acquisition of antibiotic resistance in bacteria can essentially follow two different routes: a) through chromosomal mutation and b) by the acquisition of specific resistance mechanisms through horizontal gene transfer (Figure 3). These two processes are fundamentally different in several key respects. Chromosomal mutations are the alteration of genes encoded in bacteria which lead to a phenotype of antibiotic resistance. This is a powerful adaptive mechanism, given the high constitutive mutation rate in bacteria of constituent mutations in bacteria (which is further increased in the presence of some antibiotics) and the large size of bacterial populations (which increases the chances of at least one specimen in the population acquiring the appropriate mutation).
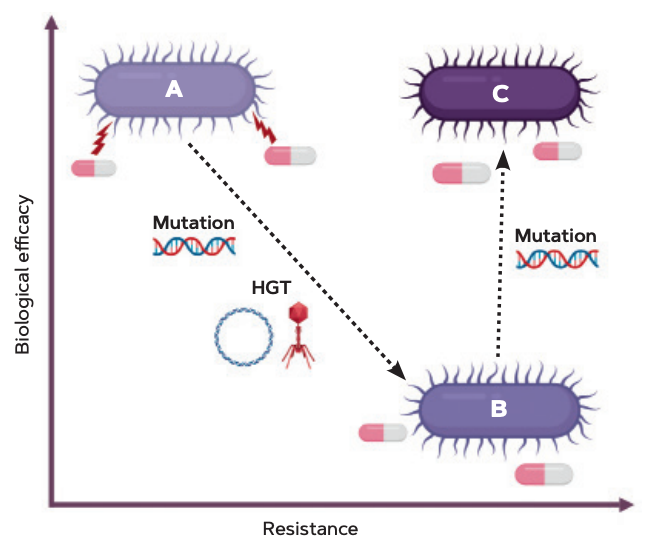
Figure 2. Diagram of the evolution of bacterial antibiotic resistance. An antibiotic-sensitive bacterium (A) evolves a resistance thanks to chromosomal mutations or horizontal gene transfer (HGT). The resistance mechanism imposes a biological cost or a reduction in biological efficacy (B), which is corrected through compensatory mutations (C). / Alvaro San Millán
Resistance-related mutations can affect the cellular targets of antibiotics and modify them so that so they are not longer recognised by antibiotics. For instance, if the action of an antibiotic is based on blocking a bacterial enzyme, any mutation altering the structure of that enzyme and therefore preventing it from being recognised by the antibiotic (ideally without significantly altering its activity) will lead to resistance. Other types of resistance-conferring mutations are those that reduce the intracellular concentration of antibiotics by their expulsion or reducing their uptake. This category includes, for example, mutations that induce the overexpression of efflux pumps that actively remove the antibiotic from the bacterial intracellular compartment. Crucially, the acquisition of resistance through chromosomal mutation is based on the alteration of the bacterium’s pre-existing genetic material and are transmitted vertically to the cell’s offspring.
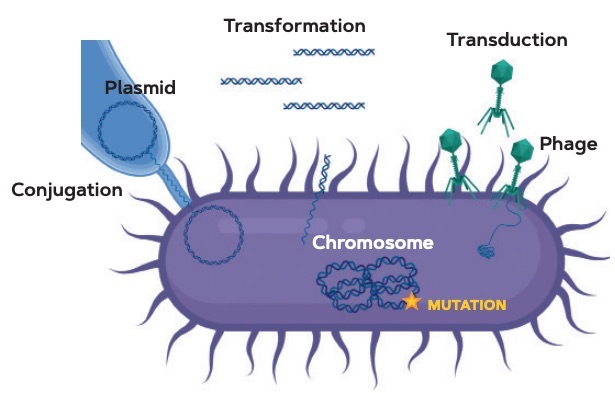
Figure 3. Diagram of the bacterial antibiotic resistance acquisition mechanisms. As shown in Figure 1, bacteria can become resistant to antibiotics through chromosomal mutations or horizontal gene transfer (HGT). The picture shows the main HGT pathways: conjugation (direct plasmid transfer between bacteria), transformation (DNA capture from the environment), and transduction (bacteriophage-mediated DNA transfer). / Álvaro San Millán
The second route to the acquisition of antibiotic resistance is horizontal gene transfer (HGT). HGT is the movement of genetic material between organisms independently of vertical inheritance and is, effectively, the direct exchange of DNA between individuals. This process plays an essential role in the evolution of living beings and is particularly frequent and relevant in bacteria compensating for their lack of sexual reproduction. HGT in bacteria can occur directly through the capture of naked DNA from the environment, which is then recombined with the bacterium’s genetic material in a process known as transformation. What is even more relevant for the evolution of innovations in bacteria is HGT transmitted by mobile genetic elements (MGEs), genetic structures capable of transferring between bacteria. Among these elements are bacteriophages (or phages) and plasmids, which mediate the processes known as transduction and conjugation, respectively (Figure 3). MGEs carry the genes necessary for their own biological cycle, but they also encode multiple accessory genes with a high adaptive value for bacteria. Thus, MGEs provide bacteria with a range of completely new genes, which allow them to adapt to new ecological niches and resist different environmental insults almost immediately. Can the reader imagine being able to acquire the eye colour of the person who is sitting next to you, the sense of smell of a dog, or even bird wings in a matter of minutes? That is the power of HGT in bacteria.

Figure 4. In addition to research into new antibiotics, we must promote their more rational use to decrease selective pressure on bacterial populations. Thus, governments around the world are releasing awareness campaigns both for their citizens and medical staff, with the aim of ending the abuse of these magic bullets. The picture shows a campaign poster for the Spanish National Plan against Antibiotic Resistance, which has been active since 2014 and whose second phase (2019–2021) has just started. / Ministerio de Sanidad, Servicios Sociales e Igualdad. Gobierno de España
Plasmids are the most relevant MGEs in the evolution of bacterial antibiotic resistance in clinical environments (San Millán, 2018). Plasmids are small circular DNA molecules that replicate independently from the bacterial chromosome and are transferred via a protein tunnel which directly connects bacteria in a process known as conjugation. These genetic elements encode sophisticated resistance mechanisms, many of them acquired from environmental bacteria that have been producing the antibiotics we currently use in hospitals for millions of years. Even more worryingly: these plasmids usually carry not one, but several different resistance mechanisms, providing resistance to multiple antibiotics in a single conjugation event. In addition, multiple copies of plasmids are kept within host bacteria, providing an increase in the dose of the genes they encode and which serves as fuel for the continuous evolution of innovations (Rodríguez-Beltrán et al., 2018; San Millán, Escudero, Gifford, Mazel, & MacLean, 2016). For all these reasons, plasmid-mediated antibiotic resistance has emerged as one of the most difficult-to-solve problems in clinical environments over the last few decades.
Conclusion
Study of the evolution of antibiotic resistance allows us to understand some of the rules that govern the evolution of innovations in bacteria and, by extension, in the rest of living beings. The evolution of resistance is testament to the incredible adaptive ability of bacteria, which normally find not one, but several genetic solutions to survive antibiotics. Therefore, if we want to continue enjoying the enormous benefits of these magic bullets in the future, we must promote the more rational use of antibiotics that will contribute to reducing the selective pressure put on bacterial populations (Figure 4). Finally, the most fascinating element in the subject of this text is probably the fact that during the minutes it took the readers to go through it, all the genetic adaptation mechanisms we have described here will have actually occurred within their own digestive tract, inside some of the approximately thirty billion bacteria inhabiting it.
REFERENCES
Buckling, A., Craig Maclean, R., Brockhurst, M. A., & Colegrave, N. (2009). The Beagle in a bottle. Nature, 457, 824–829. doi: 10.1038/nature07892
CDC. (2013). Antibiotic resistance threats in the United States. Centres for Disease Control and Prevention, US Department of Health and Human Services. Retrieved from http://www.cdc.gov/drugresistance/threat-report-2013
ECDC. (2013). Annual epidemiological report 2012. Reporting on 2010 surveillance data and 2011 epidemic intelligence data. Stockholm: ECDC. Retrieved from https://ecdc.europa.eu/en/publications-data/annual-epidemiological-report-2012-2010-data
Fleming, A. (1945, 11 December). Penicillin. Nobel Lecture. NobelPrize.org. Retrieved from https://www.nobelprize.org/uploads/2018/06/fleming-lecture.pdf
Jonas, O., Parry, I., Chisholm, D., Banatvala, N., & Ladminarayan, R. (2014). Global health threats of the 21st century. Finance and Development, 51(4), 16–20. Retrieved from https://www.imf.org/external/pubs/ft/fandd/2014/12/jonas.htm
Mayr, E. (1963). Animal species and evolution. Cambridge, MA: Harvard University Press.
O’Neill, J. (2016). Tackling drug-resistant infections globally: Final report and recommendations. London: Review on Antimicrobal Resistance.
Rodríguez-Beltrán, J., Hernández-Beltrán, J. C. R., DelaFuente, J., Escudero, J. A., Fuentes-Hernández, A., MacLean, R. C., … San Millán, A. (2018.) Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nature Ecology & Evolution, 2, 873–881. doi: 10.1038/s41559-018-0529-z
San Millán, A. (2018). Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends in Microbiology, 26(12), 978–985. doi: 10.1016/j.tim.2018.06.007
San Millán, A., Escudero, J. A., Gifford, D. R., Mazel, D., & MacLean, R. C. (2016). Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nature Ecology & Evolution, 1, 0010. doi: 10.1038/s41559-016-0010
Tan, S. Y., Grimes, S. (2010). Paul Ehrlich (1854-1915): Man with the magic bullet. Singapore Medical Journal, 51(11), 842–843.
Wagner, A. (2015). Arrival of the fittest: How nature innovates. New York: Current.
WHO. (2016). United Nations high-level meeting on antimicrobial resistance. Retrieved from https://www.who.int/antimicrobial-resistance/events/UNGA-meeting-amr-sept2016/en/
WHO. (2018). Antibiotic Resistance. Retrieved from https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance