DNA rewriting our memory
Recovering missing people through their genetic profile

Continuous advances in DNA analysis for forensic purposes have set milestones in the search for genetic identity in criminal cases, disasters, and disappearances. Technological advances in the study of our genome now allow us to infer whose remains have been found, for example, at a mass grave or an anonymous tomb, and to extrapolate where they lived, their physical appearance, or their family origin. Thanks to a series of fixed variations between individuals, the analysis of DNA of forensic interest allows the identification of individuals via their genetic profile. This identification can be carried out by comparing the profile of the human remains with those of known profiles or by their compatibility with DNA inherited by their relatives.
Keywords: missing persons, genetic profile, methylation profile, phenotypic profile, biogeographic profile.
Recovering the past
Since the discovery of genetic fingerprinting by Alec Jeffreys’ team at the University of Leicester in 1985, continuous progress has been made in the analysis of DNA for forensic purposes, both at the technological level and in terms of the type of genetic markers that can be analysed, as well as their standardisation. Thanks to a lot of research and the efforts of many groups, all under the umbrella of the International Society for Forensic Genetics (Bär et al., 1997; Bodner et al., 2016; Carracedo et al., 2000; Mundorff, Bartelink, & Mar-Cash, 2009; Parson et al., 2016), these advances have been steadily marking milestones in the search for genetic identity in order to clarify criminal causes, manage major catastrophes (Prinz et al., 2007), or investigate disappearances.
«Technological advances in the study of our genome allow us to infer to whom a given set of remains belonged»
DNA cannot be seen with the naked eye, which is why it went unnoticed for so many years. But it can always be read, and with ever sharper clarity, in human remains which may have been forgotten by family and cultural transmission, but whose history we can remember or complete through genetic fingerprints. Technological advances in the study of our genome allow us to infer to whom a given set of remains belonged, where they lived, their physical appearance, or their genealogy (Figure 1). Moreover, with the advance of genomics it is likely that we will be able to deduce many other aspects of social identity that we can still only imagine today.
We know that there is no history without memory: that is why there is always a record of success and victory; but we must also strive to reconstruct our species’ failures and shame. We can already read our Neanderthal genomic prehistory (Green et al., 2010; Noonan et al., 2006), as well as that of other ancestors that became extinct thousands of years ago – a dormant prehistory written in our own DNA (Dannemann & Kelso, 2017). Soon we will be able to recover the memory of people whose remains were forgotten or concealed in the last century, be it during war or post-war periods. In the same way, we will soon be able to rescue those who are currently being forgotten, victims of hunger, discrimination, and institutionalised injustice as well as the many ongoing conflicts and waves of migrants that these struggles generate.
Technology continues to advance, and the handling of study materials during the process of identifying the remains of loved ones exhumed from mass graves or anonymous tombs – or those recovered from Mediterranean beaches – is also being refined. Similarly, genomic sequence extraction, restoration, and interpretation techniques are also improving every day. Thus, we can now perform kinship analysis or compare the results with unambiguous samples from a victim. Sometimes we can even obtain details about some of their phenotypic traits or information about the origin of their ancestors and establish a probabilistic approximation of their age or some characteristics about their lifestyle. However, we would not want to give the impression that identifying human remains is very simple. It is not always possible to put a name and surname to these remains based on the indirect information provided by scientific study. This is because the material is often so degraded that it cannot tell us what we truly hope to discover: their individual identity.
On the walls of a city one could read «there is no learning without memory»: those who remember make an investment in the future, on preserving their identity and rigour, which will allow them to survive the famous test of time. Without memory we cannot treasure knowledge or learn from the successes and errors of past eras via their footprints or the remains that narrate the unknown story of what happened. A good account of history has to be the sum of many truths: those of heroes, but also those of anonymous people or unknown witnesses. These individuals must be given an identity so that we can learn about the time in which they lived, dreamed, or disappeared. Historians, artists, palaeontologists, anthropologists and, for a few decades now, geneticists have been involved in this work.
Given these reflections, the following text will try to explain, thanks to technological advances, what questions we can ask about these remains, which ones we can quickly answer, and which will take longer for us to discover the truth.
Looking at the present
Since the complete sequencing of the human genome at the beginning of the twenty-first century (Venter et al., 2001) multiple techniques have been developed to improve the analysis of DNA of forensic interest i.e., that used to assist in the identification of the remains of missing persons from armed conflicts, disasters, human trafficking, or other origins. DNA can be «typed» thanks to the understanding of genetic variation between humans: our genome presents a series of fixed variations between subjects that allow them to be individually identified. Thus, human remains can be identified by comparing their profiles to known profiles or by their compatibility with the genetic inheritance of identified family members. This is also the case when we carry out the most basic of forensic investigations: the biological investigation of paternity.
In this case, we compare the genetic profile of the child with that of the mother (of which there is usually no doubt because birth is a documented event). The genetic variation between the two genomes, i.e., the traits not inherited by the child from the biological mother, must necessarily come from the male parent or biological father. This type of method is also applied in missing persons cases, in which the genetic information of the most suitable relatives is compared with that of the missing person (either alive or from their remains). We look at the inheritance of this information in a «neutral» way – i.e., in this circumstance, we doubt both parents – because we are not sure whether or not those remains belong to the missing relative.
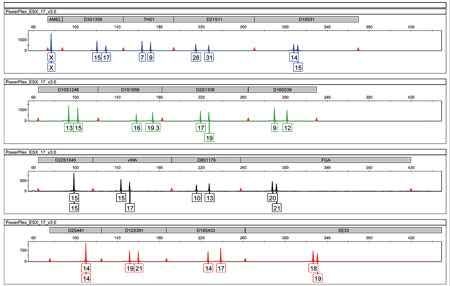
Figure 2. A genetic profile created from STR-type markers (short tandem repeats) is like a bar code which is exclusive to each person. The image shows a profile of this type obtained by analysing DNA fragments from an anonymous individual using an automatic sequencer. The colours correspond to each of the fluorescent markers used and each peak in the electropherogram represents a variant (allele) of the analysed individual. To elaborate this profile, a commercial kit was used to analyse 17 microsatellites in different chromosomes, including the amelogenin gene (the first peak shown in blue), which allows us to know whether the sex is male (XY) or, as in this case, female (XX). / Mercedes Aler
Technically, this type of identification is carried out by analysing small genomic fragments called microsatellites or STRs (short tandem repeats) which provide a specific genetic code or profile. These are repetitions of nucleotide base pairs (generally tetra- or pentanucleotides) that generate fragments of DNA of different lengths that are very polymorphic – that is, they are very variable between individuals – and that, when analysed automatically in a sequencer, provide us with a genetic profile. This is a kind of unique and exclusive bar code for each person (Figure 2), except for monozygotic identical twins who both have the same profile.
When we cannot obtain this complete profile because the initial sample is in poor condition (deteriorated remains with very fragmented or degraded DNA) or when we need more answers, we can extend the study with other types of polymorphisms which are much more abundant in our genome: SNPs (single nucleotide polymorphisms) and indel (mutations by insertion or deletion of nucleotide bases). By analysing these we increase the information available to solve many of the questions that STRs could not.
«A genetic profile is a kind of unique and exclusive bar code for each person, except for monozygotic identical twins»
There are millions of these SNP fragments in different segments of the genome, and because they are much smaller than STRs, it is more difficult for them to degrade and break up. While this analysis is still very useful, its work protocols require more processing time and, although the SNP mutation rate is lower than that of STRs, SNPs are less polymorphic and so more SNP studies must be carried out to obtain the same information obtained with STRs (Gill, 2001).
Indels, which are also considered to be a type of SNP, comprise simple biallelic insertion and deletion polymorphisms in one or a few nucleotide bases. These are simple to analyse and, like STRs, can be studied by size groups via fragment analysis. All these polymorphisms are analysed using similar methods involving the simultaneous amplification of multiple fragments (multiple reactions). These are then analysed in an automatic sequencer and stained with a fluorescent product during the polymerase chain reaction.
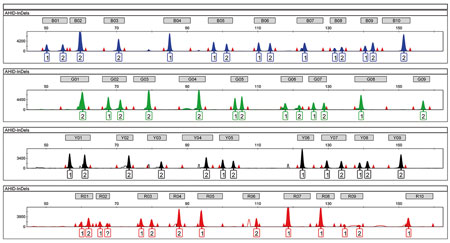
Figure 3. When the poor quality of a DNA sample makes it difficult to create a complete genetic profile, the study can be extended by analysing other polymorphisms which are abundant in our genome. These include indels – simple biallelic insertion and deletion polymorphisms comprising one or a few nucleotide bases. The picture shows a genetic profile with 38 autosomal indel markers used for identification purposes and studied by analysing DNA fragments in an automatic sequencer. Each peak in the electropherogram image represents a variant (allele) in the analysed individual. / Mercedes Aler
A specific genetic profile can be obtained for each individual based on the analysis of all the above, by analysing a different number of points in each genome: around twenty different genome points for microsatellites or STRs, around fifty for an SNP profile, and around forty for an indel profile (Figure 3). The number of analysed polymorphisms from which the genetic profile is obtained varies according to the product used for the analysis: commercial kits contain between seventeen and twenty-four and those generated by research groups include the ones that fulfil their specific project objective (for example, the elaboration of a phenotypic or ancestral profile).
In recent years, the analysis of SNP variations in DNA in relation to visible physical characteristics – known as FDP, for forensic DNA phenotyping – is gaining importance. In this context, FDP could be useful for obtaining more specific information about the missing person which could then be compared with reference samples from relatives (e.g., a photograph or a description of features like eyes, skin colour, hairline shape, etc.). In other words, although the study of phenotypic trait markers alone is insufficient to identify the missing person, these markers can provide specific answers about the missing person’s physical characteristics which could help identify them. Thus, if the remains being studied match the expectations in terms of the appearance and genetics of the person we are looking for, we will be one step closer to an answer.
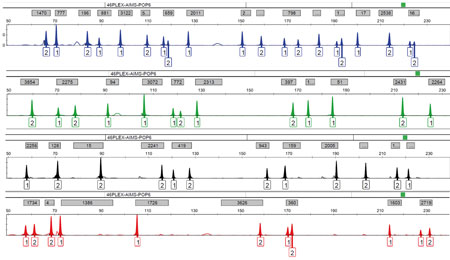
Figure 4. The analysis of SNPs (single nucleotide polymorphisms) is very useful. Because these are extremely small DNA fragments, it is more difficult for them to degrade and break down. There are also ancestry-informative markers (AIMs) which can be used to create the biogeographic profile of an individual. They are useful in the identification of unknown individuals who cannot be identified with a standard DNA profile. The image shows the genetic profile of 46 autosomal ancestry-informative indel markers analysed by studying DNA fragments in an automatic sequencer. Each peak in the electropherogram image represents a variant (allele) in the analysed individual. / Mercedes Aler
More importantly, with population-specific SNP variants (known as AIMs, or ancestry-informative markers; Figure 4) we can predict the so-called «biogeographic profile» using powerful probabilistic parameters. AIM profiles allow us to search for unknown people who are not identifiable with standard DNA profiling. This set of genetic markers makes it possible to predict ancestry on a continental and even subcontinental scale, although we must bear in mind that, in the era of globalisation and accelerated human migration in which we now live, ancestral origin does not necessarily coincide with nationality (Lipphardt, Toom, Mupepele, & Lemke, 2017).
AIM markers are already used to solve current criminal cases in which, for instance, the genetic profile of a sperm or blood stain does not allow identification if we do not already have the profile of a suspect to compare them to. In these «anonymous» profile cases, prediction of biogeographic and phenotypic data can provide answers about geographical ancestry (e.g., European, Asian, African origin, etc.), as well as the eye, hair, and skin colour of the donor of the material. Thus, the information about the features of the unknown suspect provided by AIM profiles provides useful data which can help to advance police investigations. This was the case for the 1997 criminal case of Eva Blanco in which the autosomal DNA profile and the Y chromosome of one of the suspects suggested that her brother may have perpetrated the crime. Thus, the use of these markers allowed the case to be solved almost twenty years after the crime was committed. Despite this case, investigations using AIM profiles cannot be carried out routinely in Spanish forensic laboratories because they are not within the reach of every centre, nor are they specifically regulated in Spanish legislation. When they are used, it is always under the authority of the court.
«Biogeographical profiles allow us to search for unknown people who are not identifiable with standard DNA profiling»
On the other hand, next generation sequencing (NGS) is revolutionising the forensic applications of DNA variation analysis, as is clinical genetic diagnosis, which includes the forensic applications of DNA variation analyses. The NGS technique, which is still being developed for applications in forensic cases, allows the simultaneous analysis of many different STR, SNP, and indel polymorphisms, together with the sequencing of all the internal nucleotide variants of STRs, which increases their potential for discrimination. However, the greatest advantage of NGS is that good results can still be obtained even from highly degraded material because small fragments can be analysed in great depth. In other words, NGS allows the same sample to be read multiple times, thus providing greater precision or depth when these are correlated. NGS also facilitates the analysis of methylation markers – the footprint that time leaves in DNA – to predict biological age (Abbott, 2018; Freire-Aradas et al., 2016).
Another NGS application is the distinction of monozygotic twins from biological samples. This was one of the challenges of forensic medicine, because the presumption of innocence prevails in criminal cases if it is not possible to prove which of two identical twins committed a crime. The sequencing of complete genomes with high coverage and the detection of postzygotic mutations has now made such differentiation possible (Vidaki & Kayser, 2017).
A final point worth mentioning is the key importance of using population databases and databases of forensic interest to obtain probabilistic results. The latter contain the anonymous genetic profiles of convicts and of the relatives of missing persons – «stored» for future use as potential identifiers – and are regulated by law (Ley orgánica 10/2007).
Waiting for the future
It is likely that the immediate future will bring advances in the field of epigenetics, mainly in the search for applications for the molecular changes that occur in cells as a result of environmental exposure (Vidaki & Kayser, 2017). The analysis of these environment-based variations in our genome, caused by progressive DNA methylation, allows us to obtain – analysing CpG markers – «methylation profiles» which are of specific forensic interest for studying limited biological remains (using very little DNA) to ascertain the tissue type of the remains (blood, semen, saliva, etc.) and the biological age of the donor. In addition, methylation profiles can provide information about an individual’s lifestyle such as their consumption of certain substances (e.g., tobacco, alcohol, or drugs), physical activity levels, early-life diet, etc. This is because all of these factors influence the structure and appearance of the body and so, by looking for and analysing specific markers, these traits can be established.
However, it is evident that before these variations can be used, they must first be validated by clarifying the complex and variable patterns and processes that produce them. This task is currently a real challenge because of the quantitative nature of these variations. The technological barriers of working with multiple genotyping methods would also have to be overcome for simultaneous qualitative analysis of several markers to become viable. Appropriate epigenetic markers must be identified, verification sought that valid results can be obtained from small starting amounts of DNA, the scope of these data must be investigated in order to develop predictive statistical models and, last but not least, ethical and social monitoring measures must be established and the benefits and risks of this type of research must be evaluated.
We would not want to finish this article without mentioning future possibilities for the microbiome (the DNA of the microorganisms present living within another organism), which will likely soon be able to contribute data for use in personal identification as well as information about the places the individual had lived and even some of their habits. This field of study is still in its early days as far as forensic applications are concerned, but it still has great potential for exciting technical breakthroughs, although it is not itself devoid of theoretical difficulties.
Finally, there are pending ethical and legal issues (Samuel & Prainsack, 2019) regarding the application of these and future advances, linked to the right to privacy and the right of ignorance (Figure 5). Mass sequencing techniques are taking our knowledge fully into the realm of individual genomic and epigenomic intimacy, which may have consequences in the fields of health, employment, lifestyle, and exposure to certain environmental factors. In addition, the importance of evaluating and interpreting data and of adequately narrowing them down to make them useful for the intended applications must also be considered in order to avoid misinterpreting results. As noted by the International Commission on Missing Persons (Parsons, Huel, Bajunović, & Rizvić, 2019), these misinterpretations can have important social or judicial consequences in the identifications made in war zones or after major catastrophes.
As we have already noted, all these advances require appropriate regulation. However, as always, technology moves faster than the law. Therefore, we must begin to legislate so that we can adjust our use of these techniques without affecting people’s fundamental rights.
References
Abbott, A. (2018). European scientists seek ‘epigenetic clock’ to determine age of refugees. Nature, 561(7721), 15. doi: 10.1038/d41586-018-06121-w
Bär, W., Brinkmann, B., Budowle, B., Carracedo, A., Gill, P., Lincoln, P., … Olaisen, B. (1997). DNA recommendations. Further report of the DNA Commission of the ISFH regarding the use of short tandem repeat systems. International Society for Forensic Haemogenetics, 110(4), 175–176.
Bodner, M., Bastisch, I., Butler, J. M., Fimmers, R., Gill, P., Gusmão, L., … Parson, W. (2016). Recommendations of the DNA Commission of the International Society for Forensic Genetics (ISFG) on quality control of autosomal Short Tandem Repeat allele frequency databasing (STRidER). Forensic Scientific International: Genetics, 24, 97–102. doi: 10.1016/j.fsigen.2016.06.008
Carracedo, A., Bär, W., Lincoln, P., Mayr, W., Morling, N., Olaisen, B., … Wilson M. (2000). DNA commission of the International Society for Forensic Genetics: Guidelines for mitochondrial DNA typing. Forensic Scientific International, 110(2), 79–85.
Dannemann, M., & Kelso, J. (2017). The contribution of Neanderthals to phenotypic variation in modern humans. American Journal of Human Genetics, 101(4), 578–589. doi: 10.1016/j.ajhg.2017.09.010
Freire-Aradas, A., Phillips, C., Mosquera-Miguel, A., Girón-Santamaría, L., Gómez-Tato, A., Casares de Cal, M., … Lareu, M. V. (2016). Development of a methylation marker set for forensic age estimation using analysis of public methylation data and the Agena Bioscience EpiTYPER system. Forensic Science International: Genetics, 24, 65–74. doi: 10.1016/j.fsigen.2016.06.005
Green, R. E., Krause, J., Briggs, A. W., Maricic, T., Stenzel, U., Kircher, M., … Pääbo, S. (2010). A draft sequence of the Neanderthal genome. Science, 328(5979), 710–722. doi: 10.1126/science.1188021
Gill, P. (2001). An assessment of the utility of single nucleotide polymorphisms (SNPs) for forensic purposes. International Journal of Legal Medicine, 114(4-5), 204–210.
Ley orgánica 10/2007, de 8 de octubre, reguladora de la base de datos policial sobre identificadores obtenidos a partir del ADN. (2007). Retrieved from
https://www.boe.es/eli/es/lo/2007/10/08/10/con
Lipphardt, V., Toom, V., Mupepele, A.-C., & Lemke, T. (2017). Open letter on critical approaches to forensic DNA phenotyping and biogeographical ancestry. Retrieved on 10 December 2018 from http://www.fb03.uni-frankfurt.de/67048840/Lipphardt-et-al-2017---OPEN-LETTER-ON-CRITICAL-APPROACHES-TO-FORENSIC-DNA-PHENOTYPING-AND-BIOGEOGRAPHICAL-ANCESTRY.pdf
Mundorff, A. Z., Bartelink, E. J., & Mar-Cash, E. (2009). DNA preservation in skeletal elements from the World Trade Center disaster: Recommendations for mass fatality management. Journal of Forensic Sciences, 54(4), 739–745. doi: 10.1111/j.1556-4029.2009.01045.x
Noonan, J. P., Coop, G., Kudaravalli, S., Smith, D., Krause, J., Alessi, J., … Rubin, E. M. (2006). Sequencing and analysis of Neanderthal genomic DNA. Science, 314(5802), 1113–1118. doi: 10.1126/science.1131412
Parson, W., Ballard, D., Budowle, B., Butler, J. M., Gettings, K. B., Gill, P., … Phillips, C. (2016). Massively parallel sequencing of forensic STRs: Considerations of the DNA commission of the International Society for Forensic Genetics (ISFG) on minimal nomenclature requirements. Forensic Science International: Genetics, 22, 54–63. doi: 10.1016/j.fsigen.2016.01.009
Parsons, T. J., Huel, R. M. L., Bajunović, Z., & Rizvić, A. (2019). Large scale DNA identification: The ICMP experience. Forensic Science International: Genetics, 38, 236–244. doi: 10.1016/j.fsigen.2018.11.008
Prinz, M., Carracedo, A., Mayr, W. R., Morling, N., Parsons T. J., Sajantila A., … Schneider P. M. (2007). DNA Commission of the International Society for Forensic Genetics (ISFG): Recommendations regarding the role of forensic genetics for disaster victim identification (DVI). Forensic Science International: Genetics, 1(1), 3–12. doi: 10.1016/j.fsigen.2006.10.003
Samuel, G., & Prainsack, B. (2019). Forensic DNA phenotyping in Europe: Views «on the ground» from those who have a professional stake in the technology. New Genetics and Society, 38(2), 119–141. doi: 10.1080/14636778.2018.1549984
Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., … Zhu, X. (2001). The sequence of the human genome. Science, 291(5507), 1304–1351. doi: 10.1126/science.1058040
Vidaki, A., & Kayser, M. (2017). From forensic epigenetics to forensic epigenomics: Broadening DNA investigative intelligence. Genome Biology, 18(1), 238. doi: 10.1186/s13059-017-1373-1