Can life be standardized?
Current challenges in biological standardization
The concept of standard strongly evokes machines, industries, electric or mechanical devices, vehicles, or furniture. Indeed, our technological civilization would not be possible – at least in the terms it is structured today – without universal, reliable components, whose acknowledged use results in competitive costs, robustness and interchangeability. For example, an Ikea screw can be used in a wide set of structurally dissimilar furniture and an app can be run on many different smartphones. The very concept of standardization is linked to the industrial revolution and mass production of goods through assembly lines. The question we will try to answer in the present paper is the extent to which standards and the standardization process can be accomplished in the biological realm.
Keywords: context-dependency, modularity, noise, promiscuity, synthetic biology.

The concept of standardization has traditionally been linked to universal and interchangeable elements in mechanical or electronic devices, machinery, vehicles, furniture, etc. Is it possible to apply the finitude, robustness and universality of standards to the world of molecular biology? / Marco Verch
«The fact that both living organisms and machines are subject to the laws of physics does not mean that living beings are machines»
Synthetic biology, defined as the engineering approach to biotechnology, attempts to give a systematic, standard-based flavor to genetic engineering. The central idea of synthetic biology is to make biology easier to engineer by incorporating tools and concepts borrowed from (mainly industrial and electronic) engineering. Engineering notions such as abstraction, decoupling and standardization are applied nowadays to the design of biomolecular systems. These efforts bring both new insights on the molecular intricacies and interactions in complex systems (i.e., cells and their environmental interactions), as well as advances in projects addressing a wide diversity of issues, from agriculture to medicine. Nevertheless, many critical challenges arise from this engineering approach to biotechnology, most of which are out of the scope of this article. One major issue, though, will be discussed in some detail in the next sections of this paper: the issue referring to some of the specific challenges the standardization process poses in the biological realm. The fact that both living organisms and machines are subject to the laws of physics does not mean that living beings are machines (for a recent, exhaustive work on the topic, see Nicholson, 2019). The differences between evolution-issued and human-made complex systems are linked to the difficulties for standardizing the former, as we discuss in the following sections.
Promiscuity
Every aspect of life is the outcome of networks of interactions between a diversity of components, either intracellular molecules, cells in multicellular organisms, or species in ecosystems. Thus, living beings are complex systems – in the same sense as physicists use this expression. For instance, at the molecular level, thousands of small molecules, macromolecules, and supra-macromolecular assemblies drive the ability to construct all the cell components from external feedstocks (i.e., metabolism) or the capacity to store and transmit the genetic (digital) information that must be decoded to unfold cellular functions. Proteins are key macromolecules supporting biological activities, and biochemists and structural biologists have gathered an impressive volume of details about the architecture, dynamics, and interactions of these molecular engines. Within the context of synthetic biology, proteins are usually considered interchangeable parts of the system that, through molecular interactions, can perceive and process inputs, and elicit outputs, as it occurs in any information processing system. But designed biological systems pose several issues that may result in a departure from the predicted behavior.
«Designed biological systems pose several issues that may result in a departure from the predicted behavior»
When a given protein is transplanted to a non-native context, a diversity of unintended interactions may emerge. This situation has been referred to as semantic errors in a discussion on potential failures in synthetic, reconstructed systems (Kittleson et al., 2012). For instance, the overproduction of a non-native metabolite, such as mevalonic acid, in Escherichia coli, results in toxic effects, namely, the inhibition of fatty acid biosynthesis by one of the intermediates of the heterologous pathway (Kizer et al., 2008). In addition to the native function, sculpted by natural selection, proteins often show promiscuous activities, i.e., unwanted side reactivities or interactions that in normal conditions do not represent a particular harm to the system or simply are non-adaptive. Promiscuous activities are responsible for the so-called underground metabolism (D’Ari & Casadesús, 1998), a collection of side or minor reactions, traditionally overlooked by biochemists, which are the outcome of the ability of enzymes to process substrates other than the canonical ones (Khersonsky & Tawfik, 2010). Also, molecular promiscuities have been exploited in in vitro evolution of new enzymatic activities, or even in new non-natural abilities, e.g., enzymes using nonbiological substrates or chemical elements (Arnold, 2019). In nature, promiscuous activities also may represent a drawback if they generate toxic products or inhibitors of essential functions. In this sense, there is a growing evidence of the existence of many, until recently unnoticed, metabolic repair systems that efficiently mitigate or prevent these problems (De Crécy-Lagard et al., 2018).
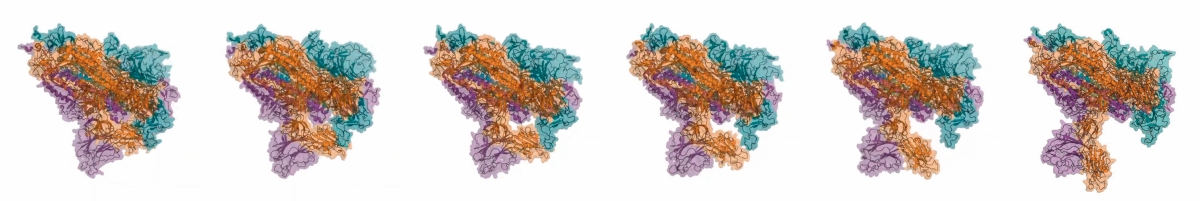
In synthetic biology, proteins are considered interchangeable parts of a system that, through molecular interactions, can perceive, process, and transmit information. However, unexpected interactions can occur in synthetic biological systems, such as promiscuous activities (i.e., reactions and interactions aside from the main function of the protein) that may lead to malfunction of the system. A demonstration of how the intrinsic flexibility of proteins reveals functions is found in the S protein of SARS-CoV-2 (in the picture): the movement of one of its structural domains exposes the binding surface to the human ACE2 protein. Therefore, one of the goals of synthetic biology must be to discover the unknown functions of proteins coded in the different genomes, and to find strategies to minimize their effects. / FoldingAtHome
At present, our fragmentary knowledge about promiscuous activities is a serious disadvantage for the de novo design of artificial circuits. Merging proteins from different evolutionary origins in a non-native context may result in the accumulation of toxic or unwanted intermediaries produced by promiscuous activities. In the future, synthetic biology may alleviate the challenge of the semantic errors (i.e., unexpected and undesirable side effects) with a joint effort in developing tools to predict and assay promiscuous activities, and in updating the public databases of enzymes and proteins. Synthetic biology may also enable a complete mapping of the metabolic «dark matter» through, for example, the discovery of the unknown function of many enzymes coded in the genomes (Ellens et al., 2017), which can lead to the design of strategies to minimize unwanted activities through protein and cellular engineering (e.g., the design of artificial supramolecular complexes or synthetic compartments that are able to channel or store the unwanted metabolites). The final outcome of all these efforts should be the accurate definition of a standard behavior of designer biomolecules acting in a non-native context, a situation that will render more predictive and optimized artificial systems. At any rate, we must remember that proteins are a case of soft matter and that promiscuity is the unavoidable outcome of their intrinsic structural flexibility and dynamic behavior. Thus, it is wise to contemplate the inevitability of promiscuity of the system components as a contingency of any synthetic biology project, evaluating its impact on the global performance of the designer system.
«Complex biological functions are the result of the joint activity of populations of entities, either molecules, cells, or tissues»
Noise
In addition to the presence of promiscuous components, we also have to consider that complex biological functions are the result of the joint activity of populations of entities, either molecules, cells, or tissues. In this sense, the stochastic behavior of the individual members of the population has been perceived as a potential difficulty for engineering biological systems, as well as an inconvenience for standardization. Even within genetically homogenous populations of cells, the stochastic behavior of individuals generates noise. This functional diversity is due to the differences and fluctuations in the available molecular machineries inside each individual cell. Even in the most ideal of the conditions, two cells will harbor different amounts of RNA polymerases (enzymes responsible of copying each gene in the corresponding template to be decoded), ribosomes (cell factories for protein synthesis), regulatory factors (that activate or inhibit other processes), and so on. Thus, when analyzed at the individual level, cells exhibit functional variability (Elowitz et al., 2002) and this phenomenon is also observed in artificially designed systems. For instance, one of the works generally considered as foundational of synthetic biology, namely, the design, simulation, and cellular implementation of an oscillatory system (the repressilator, Elowitz & Leibler, 2000), showed variability at the individual cell level due to the stochasticity of gene expression processes.
In nature, the effect of noise and of stochastic phenomena can be mitigated by the system itself, whereas in other cases those effects can be used as raw material for adaptation. In engineered biological systems, both approaches have been verified (see Kittleson et al., 2012 and references therein). In fact, standard operations of biocomputation (i.e., simple logical gates) have been successfully implemented in cells during the last two decades (Amos & Goñi-Moreno, 2018). Nevertheless, the question remains whether more complex operations could be implemented within the limits of cells, with their intrinsic noisy behaviors. Alternatively, several groups have made a virtue out of necessity by exploring biological messiness and multicellular assemblies to test the usefulness of a diversity of approaches like stochastic computation, distributed computing, and fuzzy logic. The ultimate frontier would be to transcend genetic material as the only support for logic circuits and use other levels of operation, e.g., metabolic networks, to implement more complex tasks (for a comprehensive discussion on achievements and challenges in biocomputation see Amos & Goñi-Moreno, 2018). In this context, we would require a more relaxed definition of standards.

The international iGEM synthetic biology competition is a great initiative to educate the next generations of synthetic biology researchers. Often, competition winners invent their own models instead of using the standards already validated by previous works. In the image, the room where teams of students from around the world show their projects to the rest during the competition. The picture belongs to the 2014 run. / iGEM/Justin Knight
Context dependency
There is a parallelism between the variants of an industrial model (e.g., a car) and the races or strains of animals, plants, or bacteria. The economic costs of changing the machinery, templates, and designs result in a relatively low set of variations of the most popular car models – or any other artificial device. By contrast, organisms display an amazing range of variation in the shape of virtually millions of species. For example, the favorite microbiological model organism, the bacterium Escherichia coli, has itself thousands of different strains with important genetic differences (Moradigaravand et al., 2018). This leads to the basic question on whether all the strains behave in the same way from the biotechnological point of view. In research carried out in the frame of a student competition, the iGEM contest, six E. coli strains commonly used in molecular biology were transformed with a simple device consisting of a promoter and reporter gene sequences, that is, a simple genetic circuit that yields a measurable signal usually as fluorescent light. By measuring those signals, it was possible to identify how well the circuit worked in a particular bacterial strain. Surprisingly, significant differences of expression levels were found among all six strains in five out of the six constructions (Vilanova et al., 2015). In another experiment of the same work, an E. coli strain was transformed with two almost identical gene circuits, one encoding for a red protein, and the other for a green one. Cells with the expected phenotype – those being both green and red – were in fact red, since the majority of the transformed population was either red, green, or lacked any fluorescence (Vilanova et al., 2015). These results evidence that the huge variation of strains or races in living organisms implies a lack of reproducibility in the behavior of theoretically «standard» (in the sense of «universal») genetic components. The contextual sensitivity or the influence of the genetic context on the performance of even the simplest designed circuit has been recognized as one of the challenges in synthetic biology (Kittleson et al., 2012).
«The huge variation of strains or races in living organisms implies a lack of reproducibility in the behavior of theoretically “standard” genetic components»
The iGEM competition mentioned above is a spectacular effort to educate the next generation of synthetic biology practitioners. Undergraduate students worldwide attend the contest and present a synthetic biology project which must be, at least partially, based on a cloning system called «Biobricks™». As a result of the very successful iGEM events of the last decade, Biobricks have been massively used and they are thus ideal for a meta-analysis on component reuse in synthetic biology. The analysis of the award-winning teams clearly shows, though, that most of them tend to develop their own «standards» rather than using those already available – and thus theoretically validated by previous works (Vilanova & Porcar, 2014). This leads us to the same ironical conclusion that had previously been stressed by the Nobel Prize laureate Murray Gell-Mann: «A scientist would rather use someone else’s toothbrush than another scientist’s nomenclature» (Vilanova & Porcar, 2019).
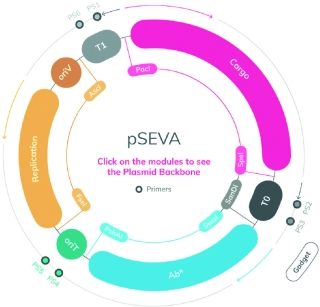
SEVA is a repository of standardized plasmids developed at the National Center for Biotechnology. In the picture, an interactive map from their 3.0 website showing the organization of the SEVA vectors. Each plasmid contains three basic modules: a cargo or DNA portion (magenta), an origin for the replication (yellow), and an antibiotic marker (blue). These plasmids can be used to build or reconstruct complex bacteria phenotypes. / SEVA/CNB-CSIC
That said, there are other examples of biological components which are arising as de facto standards in the synthetic biology community. The Standard European Vector Architecture (SEVA) platform is a series of plasmids specifically designed to be used in a range of Gram negative (now also Gram positive) species. They have been developed by the group of Víctor de Lorenzo at the National Centre for Biotechnology (Spanish National Research Council) and are provided free of charge; a total of 2,019 plasmids have been delivered to 35 countries and they have received 88 and 277 citations for the SEVA 2.0 and SEVA 1.0 version, respectively (the SEVA 3.0 has just been launched, Martínez-García et al., 2019). This highlights the importance of robustness and utility of a biological component for it to become accepted as a standard by a community of users.
Modularity
The concept of module is linked to that of standard. Mechanic devices have easily-recognizable sets of parts that are designed to be easily mounted, exchanged, or repaired. Modules, in biology, are rather obvious when it comes to anatomical structures. For example, it is not incorrect to define lungs as respiratory modules, muscles as motion, engine-like modules; or – in the case of eukaryotic cells – to consider mitochondria as internal, energy-producing modules. Indeed, endosymbionts are one of the best examples of modules in biological systems (Porcar et al., 2013). However, when we move all those well-defined and physically-contained structures to metabolic pathways (in other words, if we switch from biological hardware to biological software) the situation changes dramatically, as the degree of intercommunication, variability, noise, and enzymatic promiscuity present in metabolic pathways makes the concept of module much more blurred. This has important consequences for synthetic biology, since modularity implies a much easier design, construction, and reparation of a complex system, as it is fractioned in simpler parts. If modules do not exist, or they are too complexly connected to other modules (the latter being the case in biological systems), the goal to standardize modules/blocks/pathways becomes a huge challenge.
«Organisms display an amazing range of variation in the shape of virtually millions of species»
Conclusion
In summary, standardization in biology is an immense challenge, due to the complexity of living systems, their intrinsic variation and diversity, the promiscuity and noise in protein interactions and catalysis, the tendency of biological standards to be context-dependent, and the absence of bona fide biological modules. Solving these technical issues is just the first step towards standardization since developing standards is, above all, a social process, in which end users and policy makers agree in the choice of standards and how to use them. As we have seen in this work, we are far from having standards in biology today, but the partial successes in some cases (e.g., SEVA plasmids) and the magnitude of the benefits of having biology standardized make the endeavor worthy.
References
Amos, M., & Goñi-Moreno, A. (2018). Cellular computing and synthetic biology. In S. Stepney, S. Rasmussen, & M. Amos (Eds.), Computational Matter (pp. 93–110). Springer.
Arnold, F. H. (2019). Innovation by evolution: Bringing new chemistry to life (Nobel acceptance speech). Angewandte Chemie International Edition, 58(41), 14420–14426. http://doi.org/10.1002/anie.201907729
D’Ari, R., & Casadesús, J. (1998). Underground metabolism. BioEssays, 20(2), 181–186. http://doi.org/10.1002/(SICI)1521-1878(199802)20:2%3C181::AID-BIES10%3E3.0.CO;2-0
De Crécy-Lagard, V., Haas, D., & Hanson, A. D. (2018). Newly-discovered enzymes that function in metabolite damage-control. Current Opinion in Chemical Biology, 47, 101–108. http://doi.org/10.1016/j.cbpa.2018.09.014
Ellens, K. W., Christian, N., Singh, C., Satagopam, V. P., May, P., & Linster, C. L. (2017). Confronting the catalytic dark matter encoded by sequenced genomes. Nucleic Acids Research, 45(20), 11495–11514. http://doi.org/10.1093/nar/gkx937
Elowitz, M. B., & Leibler, S. (2000). A synthetic oscillatory network of transcriptional regulators. Nature, 403(6767), 335–338. http://doi.org/10.1038/35002125
Elowitz, M. B., Levine, A. J., Siggia, E. D., & Swain, P. S. (2002). Stochastic gene expression in a single cell. Science, 297(5584), 1183–1186. http://doi.org/10.1126/science.1070919
Khersonsky, O., & Tawfik, D. S. (2010). Enzyme promiscuity: A mechanistic and evolutionary perspective. Annual Review of Biochemistry, 79, 471–505. http://doi.org/10.1146/annurev-biochem-030409-143718
Kittleson, J. T., Wu, G. C., & Anderson, J. C. (2012). Successes and failures in modular genetic engineering. Current Opinion in Chemical Biology, 16(3-4), 329–336. http://doi.org/10.1016/j.cbpa.2012.06.009
Kizer, L., Pitera, D. J., Pfleger, B. F., & Keasling, J. D. (2008). Application of functional genomics to pathway optimization for increased isoprenoid production. Applied and Environmental Microbiology, 74(10), 3229–3241. http://doi.org/10.1128/AEM.02750-07
Martínez-García, E., Goñi-Moreno, A., Bartley, B., McLaughlin, J., Sánchez-Sampedro, L., Pascual del Pozo, H., Prieto Hernández, C., Marletta, A. S., De Lucrezia, D., Sánchez-Fernández, G., Fraile, S., & de Lorenzo, V. (2019). SEVA 3.0: An update of the Standard European Vector Architecture for enabling portability of genetic constructs among diverse bacterial hosts. Nucleic Acids Research, 48(D1), D1164–D1170. http://doi.org/10.1093/nar/gkz1024
Moradigaravand, D., Palm, M., Farewell, A., Mustonen, V., Warringer, J., & Parts, L. (2018). Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLOS Computational Biology, 14(12), e1006258. http://doi.org/10.1371/journal.pcbi.1006258
Nicholson, D. J. (2019). Is the cell really a machine? Journal of Theoretical Biology, 477, 108–126. http://doi.org/10.1016/j.jtbi.2019.06.002
Porcar, M., Latorre, A., & Moya, A. (2013). What symbionts teach us about modularity. Frontiers in Bioengineering and Biotechnology, 1, 14. http://doi.org/10.3389/fbioe.2013.00014
Vilanova, C., & Porcar, M. (2014). iGEM 2.0–refoundations for engineering biology. Nature Biotechnology, 32, 420–424. http://doi.org/10.1038/nbt.2899
Vilanova, C., & Porcar, M. (2019). Synthetic microbiology as a source of new enterprises and job creation: A Mediterranean perspective. Microbial Biotechnology, 12, 8–10. http://doi.org/10.1111/1751-7915.13326
Vilanova, C., Tanner, K., Dorado-Morales, P., Villaescusa, P., Chugani, D., Frías, A., Segredo, E., Molero, X., Fritschi, M., Morales, L., Ramón, D., Peña, C., Peretó, J., & Porcar, M. (2015). Standards not that standard. Journal of Biological Engineering, 9, 17. http://doi.org/10.1186/s13036-015-0017-9