Human uniqueness
Most cosmological views on human origins portray these as being divine, beyond the natural world. This has partly been influenced by our obvious uniqueness. Because, of all past human species –numbering perhaps over a dozen– we are the only ones to have reached historical times. If we consider ourselves remarkable it is, basically, because we are unique. But if another species such as the Neanderthal had survived, no doubt Western intellectual development would have been different.
«The most plausible explanation is that these chromosomal regions are the result of ancient hybridization with Neanderthals that has left a print on the non-african genomes of our species»
Philosophers like Plato, Aristotle, Descartes, Rousseau, Hume and Marx have debated the true meaning of human nature, but their ideas have never been assessed with the scientific method, because they lacked an empirical basis. In other cases, certain characteristics have been proposed as defining humans that have since been witnessed in other primates, or even non-primate mammals, such as self recognition in a mirror or the possession of culture or technology. These are usually cognitive and social features (which are the human characteristics we ourselves admire) that have been selected a priori because they fit into a particular ideological view of how human beings should be. This list has included the existence of language, figurative art, self-awareness, complex symbolic behaviour, the fear of death and belief in an afterlife. We can detect the subjectivity of these approaches, for example, in the case of language. The Gospel of Saint John begins dramatically with this statement: «In the beginning was the Word and the Word was with God, and the Word was God.»
Some people, including researchers in evolutionary psychology, would like to believe that this exceptional feature should be unique to our species. Obviously, nobody would be so enthusiastic about the idea proposed by the Russian writer Dostoyevsky, who said that «only humans can be so artistically cruel». But the same subjectivity also affects those who propose features that can be considered negative. Findings show, for example, that other primates are also prone to deceiving their own kind for food, sex or mother’s attention, and hence lying is not uniquely human. To see how these definitions are problematic, one has only to remember that nobody would consider someone who was dumb, non artistic, an atheist or even unconscious in hospital, to be non human. In this sense, the writer Italo Calvino was right when he said that the human race is a set of individuals defined by «belonging» or «the human race is a zone of living things that should be defined by tracing its confines». That is, it would be an approach that would be scientifically verifiable a posteriori: discover first what is common to all humans.

Marco de la Rasilla, researcher at the University of Oviedo, and Svante Pääbo, principal investigator of the Max Planck Institute, in the cave at El Sidrón in Asturias northern Spain. In addition to the fragments taken from the Vindija cave, the draft genome was completed by sequencing DNA from three Neanderthals from Mezmaiskaya (Russia), Feldhofer (Germany) and El Sidrón (Spain).
© Research team of El Sidrón
The genomics era has opened a new gateway to the study of human nature. We have available to us genomes of pongids, our closest living relatives (some of these genomes are already published, like the chimpanzee, and others, such as the bonobo and orangutan, are being analysed), and personalised human genomes (remember that the original human genome project is really a palimpsest of different individuals), such as Jim Watson and Craig Venter. However, it is possible that the former evolutionary references are too remote in time to provide highly accurate information at the level of genes with a high rate of evolutionary change. That is, those genes that have changed in the human lineage in the last tens of thousands of years, which is precisely when our species emerged. Again, the extinction of numerous hominin species in the last two million years has left a huge gap in evolutionary information that the study of the chimpanzee genome cannot bridge.
Some researchers believe that if a detailed study is made of what is shared among all human genomes, an objective definition of human nature can be built. But early data from «the 1,000 genomes» project are starting to reveal another problem: the huge variation in the genome at the intraspecific level, including features such as the duplication of entire segments of chromosomes, which had not been considered until now. In other words, we can find genetic variants present in all individuals studied but cannot be sure whether the next individual analyzed will lack them. The problem is particularly acute in more variable human populations today, those in sub-Saharan Africa, as recently shown by the publication of several genomes from Khoisan and Bantu people. A further difficulty would be to label genetic variants as «exclusive to modern humans» that could in fact have been present in other extinct hominin species. That would not help us in understanding human nature, because we are interested in precisely those variants that are present in our species alone.
Despite all these problems, it is obvious that what makes us all human is that we share common ancestors, with many interconnections that join all modern humans together in a huge evolutionary network. But that does not mean everyone shares exactly all the same genes in this process. That is, heredity flow and common evolutionary history are what bring us together as a species, not a specific marker in modern-day humans. Therefore, if we could gain access to genetic information from an evolutionary reference point closer in time than that of chimpanzees, then we would be in a position to more precisely define the evolutionary novelties unique to our species.
Neanderthal Genome Project
«Genomic researchers have found thatin several segments of the chromosome, the Neanderthal genome is more similar to the three non-African genomes than to the two African ones»
Neanderthals (Homo neanderthalensis) appear to have emerged from a series of archaic populations in the European Middle Pleistocene, often classified as Homo heidelbergensis. This was a human species that lived in Eurasia between some 400,000 and 30,000 years ago, their extinction process beginning with the arrival in Europe of our ancestors from Africa, around 40,000 years ago. The exact nature of their interactions during this process, the adaptive significance of their unique morphological features and the real extent of their cognitive abilities are still controversial among researchers today.

The author of this article, Carles Lalueza-Fox (below) is part of the multidisciplinary team studying the cave in Asturias, and has participated in the Neanderthal Genome Project.
© Research team of El Sidrón
Since 1997, molecular biology techniques have enabled the recovery of thirteen mitochondrial DNA sequences (including half a dozen of complete mitochondrial genomes or mitogenomes) of different Neanderthals including some from the Asturian archaeological site El Sidrón in northern Spain, dating back some 49,000 years. Although the mitochondrial DNA of Neanderthals differs from that of modern humans, it is highly similar among Neanderthals, even among individuals thousands of kilometres apart. The two lineages of hominins have been estimated, by molecular clock, to have split half a million years ago. All this would indicate that Neanderthals did not contribute to the mitochondrial DNA of modern humans and that they were a species with very little diversity, and therefore, of very low effective population size.
The Neanderthal Genome Project is a large scientific project led by Svante Pääbo, from the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany, conducted in collaboration with the private company 454 Life Sciences-Roche, with subsequent addition of the Solexa-Illumina company. The project began in July 2006 with the aim of drafting the sequence of the Neanderthal genome within two years. The completed draft was presented publicly on February 12th 2009, on the date commemorating Darwin’s birthday. The millions of DNA sequences took a whole year to analyse, and the corresponding publication did not see the light of day until last May in the journal Science.
«If gene flow has taken place, perhaps we should say that this is the same species seeing that the concept of biological species implies a closed reproductive unit»
This project was made possible thanks to new high throughput technology platforms with massive parallel sequencing (also known as ultrasequencing), which did not exist just four or five years ago. Such progress has enabled DNA analysis to develop from the laborious process of the past to enable the generation of complete extinct genomes, with experimental analysis becoming essentially a computerised task.
Most of the draft sequence has been carried out from three small bone samples taken from the Croatian site at Vindija, found in 1980 and labelled as Vi33.16, Vi33.25 and Vi33.26. All three are from different female individuals, although the first and last share the same mitochondrial DNA and are therefore probably related maternally. The draft sequence genome coverage is almost 2x, which means that, on average, each nucleotide in the genome (and there are some 3,100 million!) is represented by two sequences. In practice, however, this means a nucleotide may have one or two extra sequences while the neighbouring one may not have any, because the sequences are randomly distributed. So we have about 63% of the genome (one must add that the many regions formed by repeated sequences may never be known, given the original DNA fragmentation). The draft Vindija sequence has been completed with the partial sequencing of three more Neanderthals and among these we have El Sidrón 1253, from Asturias, dated to some 49,000 years ago. In the latter case, only 2.2 million nucleotides have been obtained or 0.1% of the genome.
Neither out of Africa nor multiregional, but something else
When genomic analysts began studying the Vindija genome and compared it with the complete genomes of five modern humans (two Africans belonging to the Yoruba and the San tribes, a European, a Chinese and a Papua New Guinean), they discovered something that puzzled them. And not just them, it is clear that the finding will lead to a paradigm shift regarding human evolution. I have also had to modify my previous convictions, which had been based on more limited evidence; but that’s how science works. The fact is that the results do not tie in with either of the extreme and mutually exclusive hypotheses, put forward to date, concerning the origin and evolution of our species, known as the «Out of Africa» and the Multiregional theories. It is, therefore, a new theory of human evolution.

Entrance of the Vindija cave, in Croatia. The bones used to prepare the Neanderthal draft genome were found on this site.
© Johannes Krause, Max Planck Institute for Evolutionary Anthropology
The «Out of Africa» hypothesis (sometimes known as African Eve or mitochondrial Eve) maintains that our species originated in Africa between 100,000 and 200,000 years ago and subsequently spread to the other continents, while at the same time replacing the local archaic populations (such as Neanderthals) without any interbreeding taking place. Most genetic studies, especially those based on the Y chromosome and mitochondrial DNA support this hypothesis, because the most basal branches of these uni-parental markers are always in sub-Saharan Africa, and also the diversity found is very low, and therefore recent. By contrast, the Multiregional hypothesis, advocated by certain morphologists studying fossils, argues that the human populations on each continent derived from migrations leaving Africa much earlier, around two million years ago, of very ancient hominin species such as Homo erectus or the like. These populations would have evolved locally, giving rise to different human groups in each continent, while maintaining consistent morphology and genetics (this would be essential as all human crosses yield fertile offspring) by gene flow, i.e., progressive gene exchange between one geographic area and another without great migrations, just minor crossings between adjacent populations. At the start of the twenty-first century, the vast majority of scientists and, we might add, all geneticists supported the first hypothesis and, in fact, this debate was considered closed.
Well, the genome project researchers have found that in several segments of the chromosome, the Neanderthal genome is more similar to the three non-African genomes (those from New Guinea, Europe and China) than to the two African ones. If the «Out of Africa» hypothesis were correct, we would expect the opposite, because as the African genomes are located nearer the basal branch of all mankind –and therefore closer to the trunk common to both lineages– they should be more similar to the Neanderthal genome. In fact, that is what is observed in mitochondrial DNA. The human mitochondrial DNA most similar to that of the Neanderthals is precisely that of the hypothetical African mitochondrial Eve, the «mother of all human mitochondria».
«The project results do not tie in with either of the hypotheses put forward concerning the origin and evolution of our species. It is a new theory of human evolution »
In these segments, measuring some 100,000 nucleotides in length, which are present in at least 10 of the 23 chromosomes, the evolutionary divergence between Neanderthals and non-Africans is more recent than in the rest of the genome, because the DNA sequence is almost identical. However, on comparing the same segments with those of the African genomes, these differ greatly from each other, and therefore give older divergence times. It cannot be the result of background contamination (estimated at very low levels in the Vindija samples, between 0 and 0.5%), as this would be distributed evenly throughout the genome and not in the form of discrete blocks, as found here. That is, if a sample were contaminated with foreign DNA, this would be shared equally by all chromosomes. To understand all this, imagine that we are the result of hybridization, our chromosome 16, for example, has received a copy from our father (let’s say a Neanderthal) and another from our mother (a modern human), and each copy differs in parts of its sequence. When we have offspring, we do not pass one or the other copy of our chromosome 16, but we generate a new one that is a palimpsest of the two copies we already have (via an exchange phenomenon known as genetic recombination). That is, chromosome 16 of the next generation will have a block of paternal 16, and then a block of maternal 16, etc. As we move on in time away from this hybridization point, the blocks will be fragmented by successive recombinations in the offspring, but still continue to be distinguishable in a global genomic analysis. That is what has been found in the genomes of Neanderthals and present-day non-African humans studied, and represents between 1 and 4% of the genomes of the latter.

The researchers obtained most of the DNA used for study from these bone fragments of three Neanderthal females, found in the Vindija cave in Croatia.
© Max Planck Institute for Evolutionary Anthropology
The most plausible explanation is that these chromosomal regions are the result of ancient hybridization with Neanderthals that has left traces in the genomes of our species outside of Africa. For this to have happened, the phenomenon must have occurred shortly after the common migration out of Africa, 100,000 years ago. The genomic footprint is not recent enough for the last Neanderthals to have interbred with modern humans entering Europe 40,000 years ago. Among other things, had it been so, these evolutionary traces would be found in the European individual alone, and not all three non-Africans, including the one from Oceania.
Where, then, did this amazing event take place? For many years we have known that Neanderthals and early modern humans coexisted in the area of the Middle East (and probably throughout the Middle East). In Israel, we find sites close together containing the same lithic technology from the Middle Paleolithic era but with fossils pertaining to either Neanderthals or modern humans. Datings, however, do not coincide and that had led us to believe that, despite geographical proximity, they may never have actually met. The archaic Tabun Neanderthal was dated to 120,000 years; archaic modern humans of Skhul and Qafzeh to around 90,000-100,000; and Kebara and Amud Neanderthals to around 60,000 years. Furthermore, there are other Neanderthal remains in Syria (Dederiyeh) and Iraq (Shanidar). It is likely that certain crossbreeding took place in this region when our ancestors began to leave Africa. When a colonizing population like the modern humans comes across a resident population, even a very small number of interbreeding events between the two populations could leave a very large genetic footprint on the former, given the dynamics of expansion of the colonizing population. As a result, only the gene flow from the resident to the colonizing population is usually detected, as is the case. The fact that mitochondrial lineages are clearly different can indicate a sexual bias in these crosses (male Neanderthals with modern human females), although this fact could also be consistent with the random loss of mitochondria of Neanderthal origin over the generations if gene flow was, as it seems, limited.
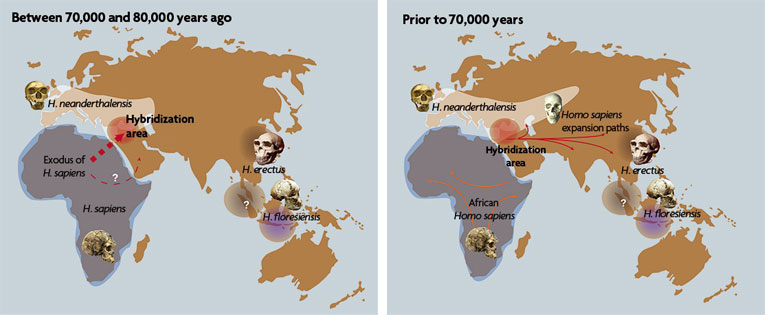
These maps show how modern humans and Neanderthals interacted between 70,000 and 80,000 years ago. According to investigations conducted by researchers at the Max Planck Institute in collaboration with Spanish scientists from the CSIC, this hybridization may have occurred in the Middle East (map on the left), with descendents spreading from there to the regions of Eurasia and Asia, as shown in the map of the right.
© CSIC Communication / Antonio Rosas
Finally, there are other regions of the Neanderthal genome that show greater evolutionary depth than the rest. Following the same reasoning, they most likely correspond to earlier traces of hybridizations with even more archaic humans. Obviously, we do not have the genome of a Homo erectus or Homo ancestor to check whether they share these genomic regions, but it would all appear to indicate that different human expansions would have led to repeated gene flow with other groups from previous population expansions.
A recently published study also points in this direction. This relates the recovery of the mitochondrial genome of a small finger bone found in the Siberian Denisova cave and dated to between about 30,000 and 48,000 years old. The mitochondrial sequence corresponds to a lineage that differs greatly from both modern humans and Neanderthals, and this divergence time has been estimated at one million years. While journalists interpreted it as the genetic finding of a new species, we must remember that there is no direct correspondence between genetic divergence and difference in species. Some species are geographically structured and that causes significant developmental differences between individuals. For example, it is known that there were two groups of mitochondrial lineages among mammoths, which diverged between 1.5 and 2 million years ago, and nobody has classified them as two different mammoth species. Could it be that genomically, morphologically and culturally Denisova was another Neanderthal, but whose mitochondrial lineage held traces of a gene flow with earlier hominins? Obviously, that can be learnt by studying its nuclear genome, as the peculiar form of transmission of mitochondrial DNA masks the possible evolutionary explanations.
An unexpected dilemma
«To understand what these changes imply, we will have to study them one by one, and understand their differential function in modern humans and Neanderthals»
Now, all this poses a new problem. To know what is uniquely human we need to have an external evolutionary point of reference, but if there is evidence of gene transfer between lineages of Neanderthals and modern humans then we may not have one. That is, if gene flow has taken place, perhaps we should say that this is the same species seeing that the concept of biological species implies a closed reproductive unit. At the genetic level, there is nothing to show that two populations (or two individuals) belong to different species –just a few genetic changes may lead to reproductive barriers–. The same goes for morphology. There are known cases –the so-called cryptic species complex– of organisms that are almost indistinguishable externally but belong to different species. Nonetheless, the concept of species remains useful; let us say it is an operational concept that biologists need to work with, from taxonomists to conservationists. Darwin himself believed that the evolutionary lineages were a continuum and division into species was somewhat arbitrary. He was right, for example, it is absurd to consider that at some point in the Neanderthal lineage we have an individual who is Neanderthal while their parents are still Homo heidelbergensis.

Several Neanderthal skulls with a Homo sapiens skull in the background. The publication of the first draft of the Neanderthal genome has revealed that Eurasian Homo sapiens share between 1% and 4% of their DNA with Neanderthals.
© CSIC Communication
Clearly, there is evidence suggesting that they would have been two different species: for example, the characteristic Neanderthal morphology falls outside the range of variation of today’s humanity, and various genetic markers, like the aforementioned mitochondrial DNA, show a clear separation between the two lineages. The most plausible thought is that there was a restricted hybridisation phenomenon that took place when the two species came into contact perhaps 100,000 years ago. This unexpected finding does not preclude, though it seems difficult, our initial approach to characterizing human nature.
The genetic definition of human beings
The most straightforward way to find singularities in the Neanderthal genome is to find those genes showing amino acid changes that, in all probability, affect the function of the encoded protein. Seventy-eight genes have been located with these types of changes, but because of the low genome coverage there could be up to three times more. Some genes on the list are easy to interpret. For example, the changes in five olfactory receptors, which are probably inactivated nowadays because they are no longer important to our survival; we, more than any other primate, are essentially visual animals.
In five other genes there are two amino acid changes (implying an even more important functional change). We have the SPAG17 gene, which appears to play an important role in the movement of sperm; the TTF1 gene, a transcription factor that acts by activating other genes; the DCHS-1 gene, which encodes for a protein involved in wound healing; the RPTN gene, which encodes a protein that is expressed in the epidermis and is involved in the sweat glands, the root hairs and tongue papillae; and the SOLH gene, which encodes a protein of unknown function.

Researcher, Svante Pääbo, with a reconstruction of a Neanderthal skull. The Max Planck Institute scientist, one of the most renowned experts on fossil DNA, has led the Neanderthal Genome Project, which published its findings last May.
© Frank Vinken
Moreover, 82 regions have been located that show signs of having been positively selected by evolutionary forces (detected because this process leaves a surrounding area of low genetic diversity). In one of these regions is the AUTS2 gene, which encodes a protein that is expressed in the brain during neural development, and which, when mutated, can cause autism. Autism is a disorder characterized by affecting social behaviour, attention span and planning and communication ability of the affected individual. Interestingly, other genes related to autism, such as ACCN1 and CADP2, and some associated with schizophrenia (NRG3) and cognitive impairment related to Down’s syndrome (DYRK1A) also appear to be under positive selection in modern humans. It has long been suggested that the remarkable human cognitive development that took place brought with it a burst of new mental disorders when mutations occurred in the genes involved in this evolutionary process. That is, our mental disorders are the result of who we are, what is now called, in military contexts «collateral damage». This evidence suggests that there may be substantial differences between us and the Neanderthals in cognitive aspects, something that should be investigated.
Other genes located in chromosomal regions with signs of selection include the THADA gene, which has been associated with type 2 diabetes. Possibly, this gene had a different function in Neanderthals and modern humans, and could thus also indicate differences in metabolism, perhaps due to differences in diet. Another interesting gene located in one of these areas is RUNX2. We know that when inactivated by mutation it leads to an ossification disorder (known as Cleidocranial dysplasia) the common features of which include delayed closure of cranial sutures, an abnormal bulging of the forehead, an undeveloped collarbone and a bell-shaped chest. Curiously, some of these features exaggerated by the disorder correspond to morphological traits that differ between us and Neanderthals, which are characterized, among other things, by presenting elongated skulls, rounded behind and with a slanting forehead.
So we have evidence that something happened to genes like SPAG17, TTF1, RPTN, SOLH, TRPM1, NRG3, DYRK1A, RUNX2, THADA, AUTS2, CADP2 and ACCN1. To this provisional list of suspects should be added dozens of other genes that show an amino acid change. This short list, which includes genes involved in the physiology of skin, pigmentation, cognition, bone development and metabolism, could be a key to defining our species. However, to understand what these changes imply, we will have to study them one by one, and understand their differential function in modern humans and Neanderthals. The easiest way to do this is to create transgenic mice with these Neanderthal genes. The «neanderthalization» of mice will be one of the scientific outcomes of the Neanderthal genome project, and will involve many years of research and possibly spectacular findings. But for the moment, we are beginning to draw up a list revealing the genetic clues underpinning humanity. Not only will we discover how Neanderthals felt, thought, ate and became ill, but we will know what makes us different. Now, 30,000 years after their extinction, our evolutionary mirror –the Neanderthals– will help us define ourselves, and we can be ourselves for the first time.
Green, R. E. et al., 2010. «A Draft Sequence of the Neanderthal genome.» Science, 328 (5979): 710-722.